Details of Host Protein-DME Interaction (HOSPPI)
| General Information of Drug-Metabolizing Enzyme (DME ID: DME0002) | |||||
|---|---|---|---|---|---|
| DME Name | Aromatase (CYP19A1), Homo sapiens | DME Info | |||
| UniProt ID | |||||
| EC Number | EC: 1.14.14.14 (Click to Show/Hide the Complete EC Tree) | ||||
| Lineage | Species: Homo sapiens (Click to Show/Hide the Complete Species Lineage) | ||||
| Interactome | |||||
| Disease Specific Interactions between Host Protein and DME (HOSPPI) | |||||
|---|---|---|---|---|---|
| ICD Disease Classification Healthy | |||||
| ICD-11: Healthy | Click to Show/Hide the Full List of HOSPPI: 8 HOSPPI | ||||
| Oligomerization | |||||
| Mutated NADPH-CYP450 reductase (mPOR) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | mPOR-CYP19A1 heterooligomerization | [2] | |||
| Studied Cell Lines | Escherichia coli BL21(DE3) cell line | ||||
| Description | Mutated NADPH-CYP450 reductase (mPOR) is reported to heterooligomerize with the CYP19A1 protein, which leads to a suppressed activity of the drug-metabolizing enzyme Aromatase. As a result, the interaction between mPOR and CYP19A1 can inhibit the drug-metabolizing process of Aromatase. | ||||
| NADPH-CYP450 reductase (POR) | Health | Heterodimer | |||
| Uniprot ID | |||||
| Interaction Name | POR-CYP19A1 heterodimerization | [3] | |||
| Studied Cell Lines | Human embryonic kidney cell line (HEK293) | ||||
| Description | NADPH-CYP450 reductase (POR) is reported to heterodimerize with the CYP19A1 protein, which leads to activation of the drug-metabolizing enzyme Aromatase. As a result, the interaction between POR and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Steroid 21-hydroxylase (CYP21A2) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP21A2-CYP19A1 heterooligomerization | [6] | |||
| Studied Cell Lines | Escherichia coli JM109 competent cell line | ||||
| Affected Substrate(s): | pregnenolone | ||||
| Description | Steroid 21-hydroxylase (CYP21A2) is reported to heterooligomerize with the CYP19A1 protein, which leads to a decreased activity of the drug-metabolizing enzyme Aromatase. As a result, the interaction between CYP21A2 and CYP19A1 can decrease the drug-metabolizing process of Aromatase. | ||||
| Transcription-factor regulation | |||||
| Nuclear receptor family 0 B1 (NR0B1) | Health | Repression | |||
| Uniprot ID | |||||
| Interaction Name | NR0B1-CYP19A1 interaction | [4], [5] | |||
| Studied Cell Lines | Human endometriotic and endometrial stromal cell lines | ||||
| Ensembl ID | |||||
| Description | Nuclear receptor family 0 B1 (NR0B1) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between NR0B1 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Steroidogenic factor 1 (NR5A1) | Health | Activation | |||
| Uniprot ID | |||||
| Interaction Name | NR5A1-CYP19A1 interaction | [4], [7] | |||
| Studied Cell Lines | Human endometriotic and endometrial stromal cell lines | ||||
| Ensembl ID | |||||
| Description | Steroidogenic factor 1 (NR5A1) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between NR5A1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Upstream stimulatory 1 (USF1) | Health | Repression | |||
| Uniprot ID | |||||
| Interaction Name | USF1-CYP19A1 interaction | [8] | |||
| Studied Cell Lines | Human trophoblast cell line | ||||
| Ensembl ID | |||||
| Description | Upstream stimulatory 1 (USF1) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between USF1 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Upstream stimulatory 2 (USF2) | Health | Repression | |||
| Uniprot ID | |||||
| Interaction Name | USF2-CYP19A1 interaction | [8] | |||
| Studied Cell Lines | Human trophoblast cell line | ||||
| Ensembl ID | |||||
| Description | Upstream stimulatory 2 (USF2) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between USF2 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-378a-3p | Health | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-378a-3p--CYP19A1 regulation | [1] | |||
| Studied Cell Lines | Granulosa cells | ||||
| Description | hsa-miR-378a-3p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
| ICD Disease Classification 02 Neoplasms | |||||
| ICD-11: 2A00 Brain cancer | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Brain neuroblastoma | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP19A1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 1.71E-06; delta-beta: -2.04E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP19A1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between DNMT and CYP19A1 can moderatly affect the drug-metabolizing process of Aromatase. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
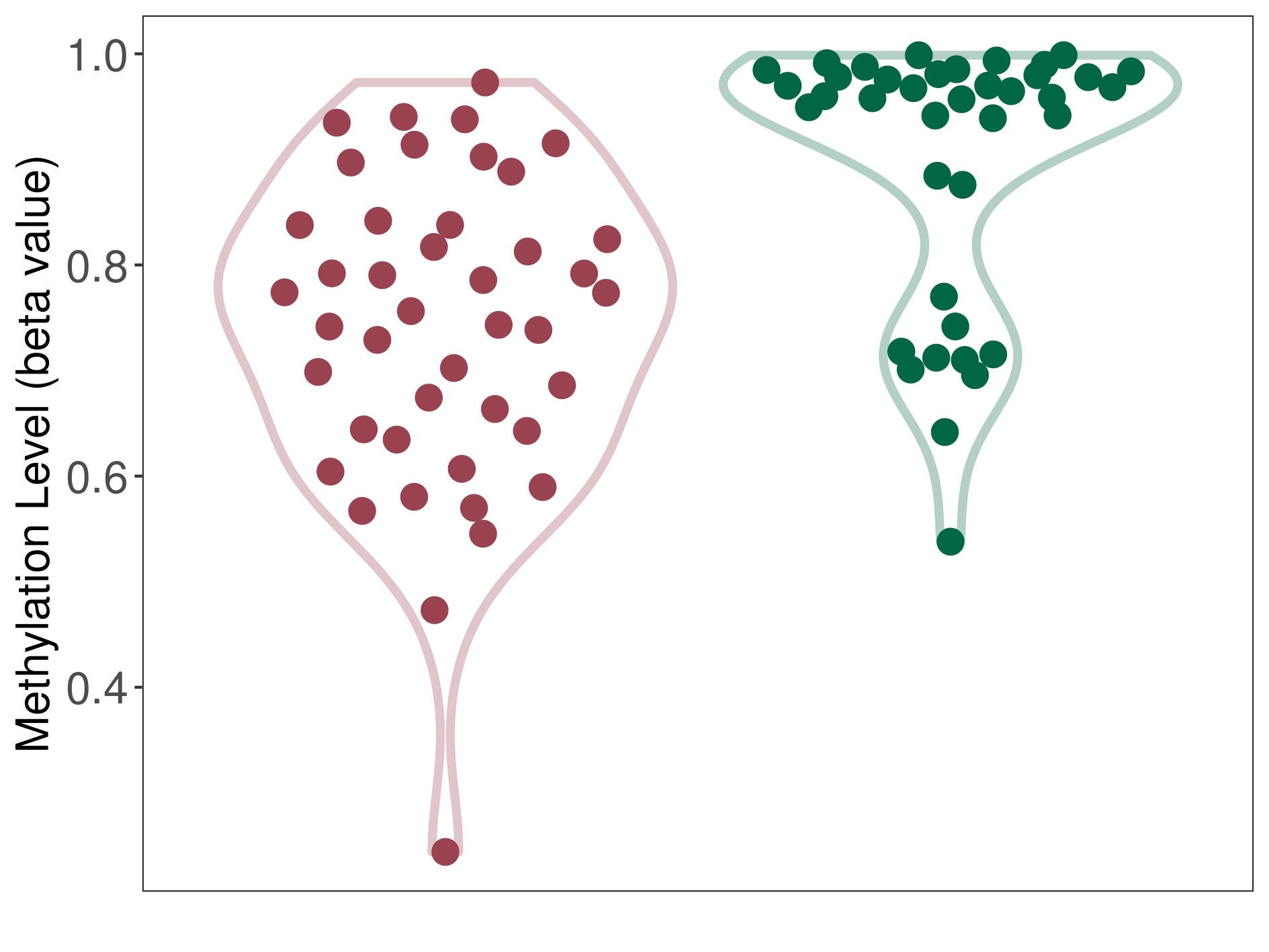
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2B30 Lymphoma | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Lymphoma | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP19A1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 1.15E-06; delta-beta: -2.09E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP19A1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between DNMT and CYP19A1 can moderatly affect the drug-metabolizing process of Aromatase. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
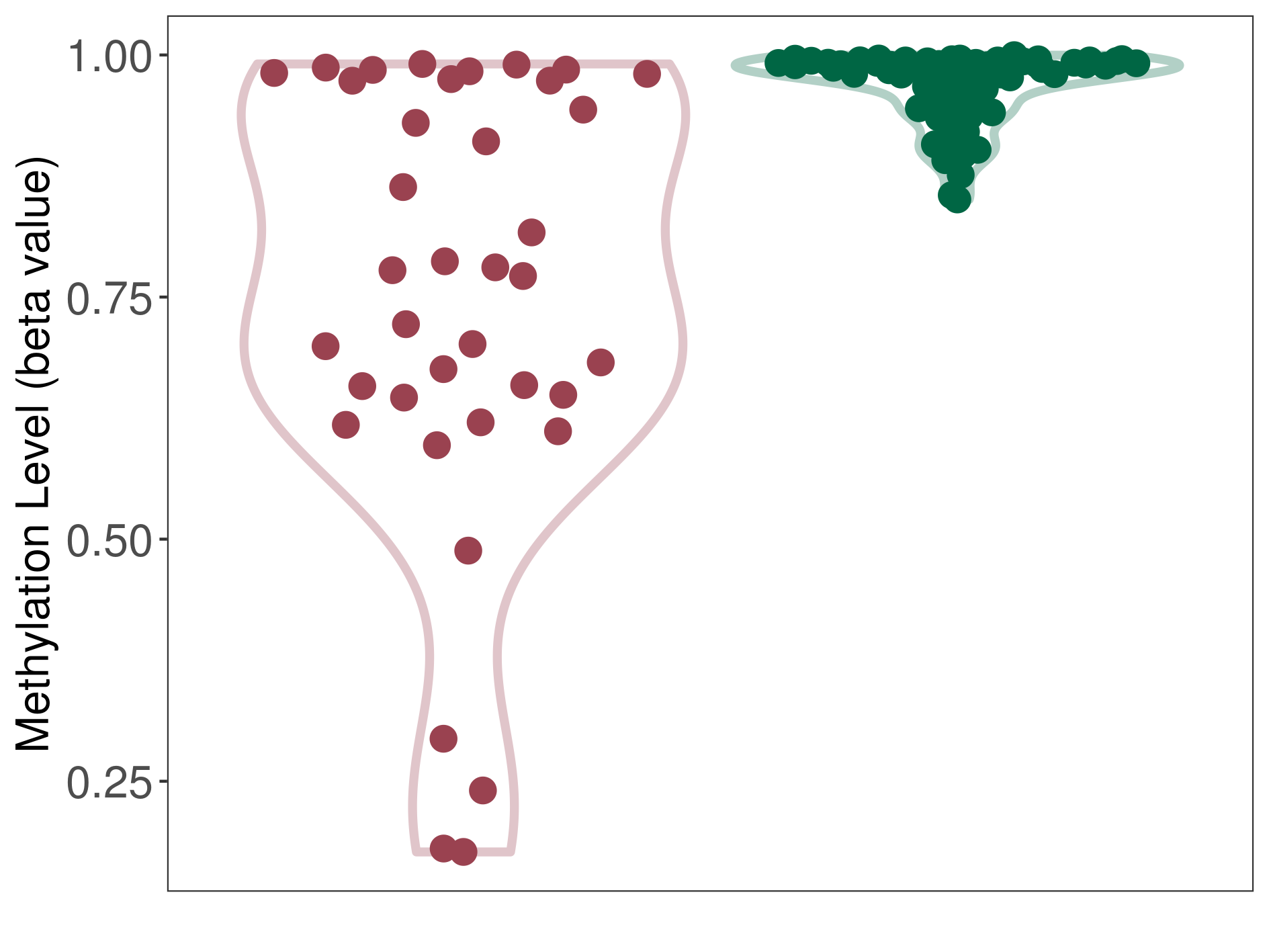
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C60 Breast cancer | Click to Show/Hide the Full List of HOSPPI: 7 HOSPPI | ||||
| Oligomerization | |||||
| Progesterone receptor 1 (PGRMC1) | Breast cancer | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | PGRMC1-CYP19A1 heterooligomerization | [16] | |||
| Studied Cell Lines | Breast carcinoma cell line (MCF7) | ||||
| Affected Substrate(s): | Androst-4-ene-3,17-dione (Metabolic product: Androst-4-ene-3,17-dione conversion) | ||||
| Description | Progesterone receptor 1 (PGRMC1) is reported to heterooligomerize with the CYP19A1 protein, which leads to activation of the drug-metabolizing enzyme Aromatase. As a result, the interaction between PGRMC1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Transcription-factor regulation | |||||
| AMP element-binding 1 (CREB1) | Breast cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | CREB1-CYP19A1 interaction | [9], [10] | |||
| Studied Cell Lines | MCF-7 and MDA-MB-231 cell lines | ||||
| Ensembl ID | |||||
| Description | AMP element-binding 1 (CREB1) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between CREB1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Estrogen receptor (ESR1) | Breast cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | ESR1-CYP19A1 interaction | [11] | |||
| Studied Cell Lines | SK-BR-3 and MCF-7 cell lines | ||||
| Ensembl ID | |||||
| Description | Estrogen receptor (ESR1) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between ESR1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Estrogen receptor-like 1 (ESRRA) | Breast cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | ESRRA-CYP19A1 interaction | [10], [12], [13] | |||
| Studied Cell Lines | MCF-7 and MDA-MB-231 cell lines | ||||
| Ensembl ID | |||||
| Description | Estrogen receptor-like 1 (ESRRA) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between ESRRA and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Glucocorticoid receptor (NR3C1) | Breast cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | NR3C1-CYP19A1 interaction | [9] | |||
| Studied Cell Lines | MCF-7 cell line | ||||
| Ensembl ID | |||||
| Description | Glucocorticoid receptor (NR3C1) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between NR3C1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Progesterone receptor (PGR) | Breast cancer | Repression | |||
| Uniprot ID | |||||
| Interaction Name | PGR-CYP19A1 interaction | [15] | |||
| Studied Cell Lines | T47D and MCF-7 cell lines | ||||
| Ensembl ID | |||||
| Description | Progesterone receptor (PGR) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between PGR and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Non-coding RNA regulation | |||||
| hsa-let-7f-5p | Breast cancer | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-let-7f-5p--CYP19A1 regulation | [14] | |||
| Studied Cell Lines | MCF-7 and SK-BR-3 cell lines | ||||
| Description | hsa-let-7f-5p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
| ICD-11: 2C73 Ovarian cancer | Click to Show/Hide the Full List of HOSPPI: 3 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Nuclear factor kappa-B p105 (NFKB1) | Ovarian cancer | Repression | |||
| Uniprot ID | |||||
| Interaction Name | NFKB1-CYP19A1 interaction | [17] | |||
| Studied Cell Lines | KGN cell line | ||||
| Ensembl ID | |||||
| Description | Nuclear factor kappa-B p105 (NFKB1) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between NFKB1 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| RING finger protein 53 (BRCA1) | Ovarian cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | BRCA1-CYP19A1 interaction | [7] | |||
| Studied Cell Lines | KGN cell line | ||||
| Description | RING finger protein 53 (BRCA1) is reported to activate the transcription of CYP19A1 gene, which leads to an increased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between BRCA1 and CYP19A1 can activate the drug-metabolizing process of Aromatase. | ||||
| Transcription factor p65 (RELA) | Ovarian cancer | Repression | |||
| Uniprot ID | |||||
| Interaction Name | RELA-CYP19A1 interaction | [17], [18] | |||
| Studied Cell Lines | KGN cell line | ||||
| Ensembl ID | |||||
| Description | Transcription factor p65 (RELA) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between RELA and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| ICD-11: 2C75 Placenta cancer | Click to Show/Hide the Full List of HOSPPI: 2 HOSPPI | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-106a-5p | Choriocarcinoma | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-106a-5p--CYP19A1 regulation | [19] | |||
| Studied Cell Lines | Human choriocarcinoma JEG-3 cell line | ||||
| Description | hsa-miR-106a-5p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
| hsa-miR-19b-3p | Choriocarcinoma | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-19b-3p--CYP19A1 regulation | [19] | |||
| Studied Cell Lines | Human choriocarcinoma JEG-3 cell line | ||||
| Description | hsa-miR-19b-3p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
| ICD-11: 2D11 Adrenocortical carcinoma | Click to Show/Hide the Full List of HOSPPI: 2 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Steroidogenic factor 1 (NR5A1) | Adrenocortical carcinoma | Repression | |||
| Uniprot ID | |||||
| Interaction Name | NR5A1-CYP19A1 interaction | [5] | |||
| Studied Cell Lines | Adrenocortical carcinoma cell line | ||||
| Ensembl ID | |||||
| Description | Steroidogenic factor 1 (NR5A1) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between NR5A1 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-98-5p | Adrenocortical carcinoma | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-98-5p--CYP19A1 regulation | [20] | |||
| Studied Cell Lines | H295R cell line | ||||
| Description | hsa-miR-98-5p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
| ICD-11: 2F32 Benign ovarian neoplasm | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Transcription factor AP-1 (JUN) | Virilizing Brenner Tumor | Repression | |||
| Uniprot ID | |||||
| Interaction Name | JUN-CYP19A1 interaction | [21] | |||
| Studied Cell Lines | Fibrothecoma-like and luteinized stromal cell line | ||||
| Ensembl ID | |||||
| Description | Transcription factor AP-1 (JUN) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between JUN and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| ICD Disease Classification 16 Genitourinary system diseases | |||||
| ICD-11: GA10 Endometriosis | Click to Show/Hide the Full List of HOSPPI: 3 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Liver activator protein (LAP) | Endometriosis | Repression | |||
| Uniprot ID | |||||
| Interaction Name | LAP-CYP19A1 interaction | [22] | |||
| Studied Cell Lines | Human endometriotic stromal cell line | ||||
| Ensembl ID | |||||
| Description | Liver activator protein (LAP) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between LAP and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Nuclear factor NF-IL6-beta (CEBPD) | Endometriosis | Repression | |||
| Uniprot ID | |||||
| Interaction Name | CEBPD-CYP19A1 interaction | [22] | |||
| Studied Cell Lines | Human endometriotic stromal cell line | ||||
| Ensembl ID | |||||
| Description | Nuclear factor NF-IL6-beta (CEBPD) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between CEBPD and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| Wilms tumor protein (WT1) | Endometriosis | Repression | |||
| Uniprot ID | |||||
| Interaction Name | WT1-CYP19A1 interaction | [4] | |||
| Studied Cell Lines | Human endometriotic stromal cell line | ||||
| Ensembl ID | |||||
| Description | Wilms tumor protein (WT1) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between WT1 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| ICD-11: GA30 Menopausal disorder | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Forkhead box protein L2 (FOXL2) | Menopausal disorder | Repression | |||
| Uniprot ID | |||||
| Interaction Name | FOXL2-CYP19A1 interaction | [23] | |||
| Studied Cell Lines | CHO cell line | ||||
| Ensembl ID | |||||
| Description | Forkhead box protein L2 (FOXL2) is reported to repress the transcription of CYP19A1 gene, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. As a result, the interaction between FOXL2 and CYP19A1 can repress the drug-metabolizing process of Aromatase. | ||||
| ICD Disease Classification 18 Pregnancy/childbirth/the puerperium | |||||
| ICD-11: JA24 Preeclampsia | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-515-5p | Preeclampsia | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-515-5p--CYP19A1 regulation | [24] | |||
| Studied Cell Lines | Preeclampsia cells | ||||
| Description | hsa-miR-515-5p is reported to suppress CYP19A1 mRNA translation by binding to the 3' untranslated region (3'UTR) of CYP19A1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Aromatase. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

