Details of Host Protein-DME Interaction (HOSPPI)
| General Information of Drug-Metabolizing Enzyme (DME ID: DME0013) | |||||
|---|---|---|---|---|---|
| DME Name | Cytochrome P450 2E1 (CYP2E1), Homo sapiens | DME Info | |||
| UniProt ID | |||||
| EC Number | EC: 1.14.14.1 (Click to Show/Hide the Complete EC Tree) | ||||
| Lineage | Species: Homo sapiens (Click to Show/Hide the Complete Species Lineage) | ||||
| Interactome | |||||
| Disease Specific Interactions between Host Protein and DME (HOSPPI) | |||||
|---|---|---|---|---|---|
| ICD Disease Classification Healthy | |||||
| ICD-11: Healthy | Click to Show/Hide the Full List of HOSPPI: 7 HOSPPI | ||||
| Oligomerization | |||||
| Cytochrome b5 (CYB5A) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYB5A-CYP2E1 heterooligomerization | [1], [2] | |||
| Studied Cell Lines | Escherichia coli cell line | ||||
| Affected Substrate(s): | Chlorzoxazone (Metabolic product: Chlorzoxazone 6-hydroxylation) | ||||
| Activity | Vmax = 9.0 nmol/min/nmol P450 | ||||
| Description | Cytochrome b5 (CYB5A) is reported to heterooligomerize with the CYP2E1 protein, which leads to an increased activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between CYB5A and CYP2E1 can facilitate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Cytochrome P450 2A6 (CYP2A6) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP2E1, POR and CYP2A6 heterooligomerization | [3] | |||
| Studied Cell Lines | Baculovirus expression system | ||||
| Affected Substrate(s): | Coumarin N-nitrosodimethylamine | ||||
| Description | Cytochrome P450 2A6 (CYP2A6) and NADPH-CYP450 reductase (POR) are reported to heterooligomerize with the CYP2E1 protein, which leads to a decreased activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction among CYP2A6, POR and CYP2E1 can decrease the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Cytochrome P450 2A6 (CYP2A6) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP2A6-CYP2E1 heterooligomerization | [4] | |||
| Studied Cell Lines | Living cells | ||||
| Description | Cytochrome P450 2A6 (CYP2A6) is reported to heterooligomerize with the CYP2E1 protein, which leads to a suppressed activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between CYP2A6 and CYP2E1 can inhibit the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Cytochrome P450 2D6 (CYP2D6) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP2D6-CYP2E1 heterooligomerization | [5] | |||
| Studied Cell Lines | Insect cell microsomes | ||||
| Affected Substrate(s): | 7-Methoxy-4-(trifluoromethyl) coumarin (Metabolic product: 7-MFC O-demethylation) | ||||
| Description | Cytochrome P450 2D6 (CYP2D6) is reported to heterooligomerize with the CYP2E1 protein, which leads to a decreased activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between CYP2D6 and CYP2E1 can decrease the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Cytochrome P450 3A4 (CYP3A4) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP3A4-CYP2E1 heterooligomerization | [6] | |||
| Studied Cell Lines | Insect cell microsomes | ||||
| Affected Substrate(s): | 7-Methoxy-4-(trifluoromethyl) coumarin (Metabolic product: 7-MFC O-demethylation) | ||||
| Description | Cytochrome P450 3A4 (CYP3A4) is reported to heterooligomerize with the CYP2E1 protein, which leads to activation of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between CYP3A4 and CYP2E1 can activate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Cytochrome P450 3A5 (CYP3A5) | Health | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | CYP3A5-CYP2E1 heterooligomerization | [6] | |||
| Studied Cell Lines | Insect cell microsomes | ||||
| Affected Substrate(s): | 7-Methoxy-4-(trifluoromethyl) coumarin (Metabolic product: 7-MFC O-demethylation) | ||||
| Description | Cytochrome P450 3A5 (CYP3A5) is reported to heterooligomerize with the CYP2E1 protein, which leads to activation of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between CYP3A5 and CYP2E1 can activate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| NADPH-CYP450 reductase (POR) | Health | Heterodimer | |||
| Uniprot ID | |||||
| Interaction Name | POR-CYP2E1 heterodimerization | [3], [7] | |||
| Studied Cell Lines | Baculovirus expression system | ||||
| Affected Substrate(s): | N-nitrosodimethylamine | ||||
| Description | NADPH-CYP450 reductase (POR) is reported to heterodimerize with the CYP2E1 protein, which leads to an altered activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between POR and CYP2E1 can modulate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| ICD Disease Classification 02 Neoplasms | |||||
| ICD-11: 2A00 Brain cancer | Click to Show/Hide the Full List of HOSPPI: 8 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Medulloblastoma | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 1.75E-60; delta-beta: -2.61E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP2E1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
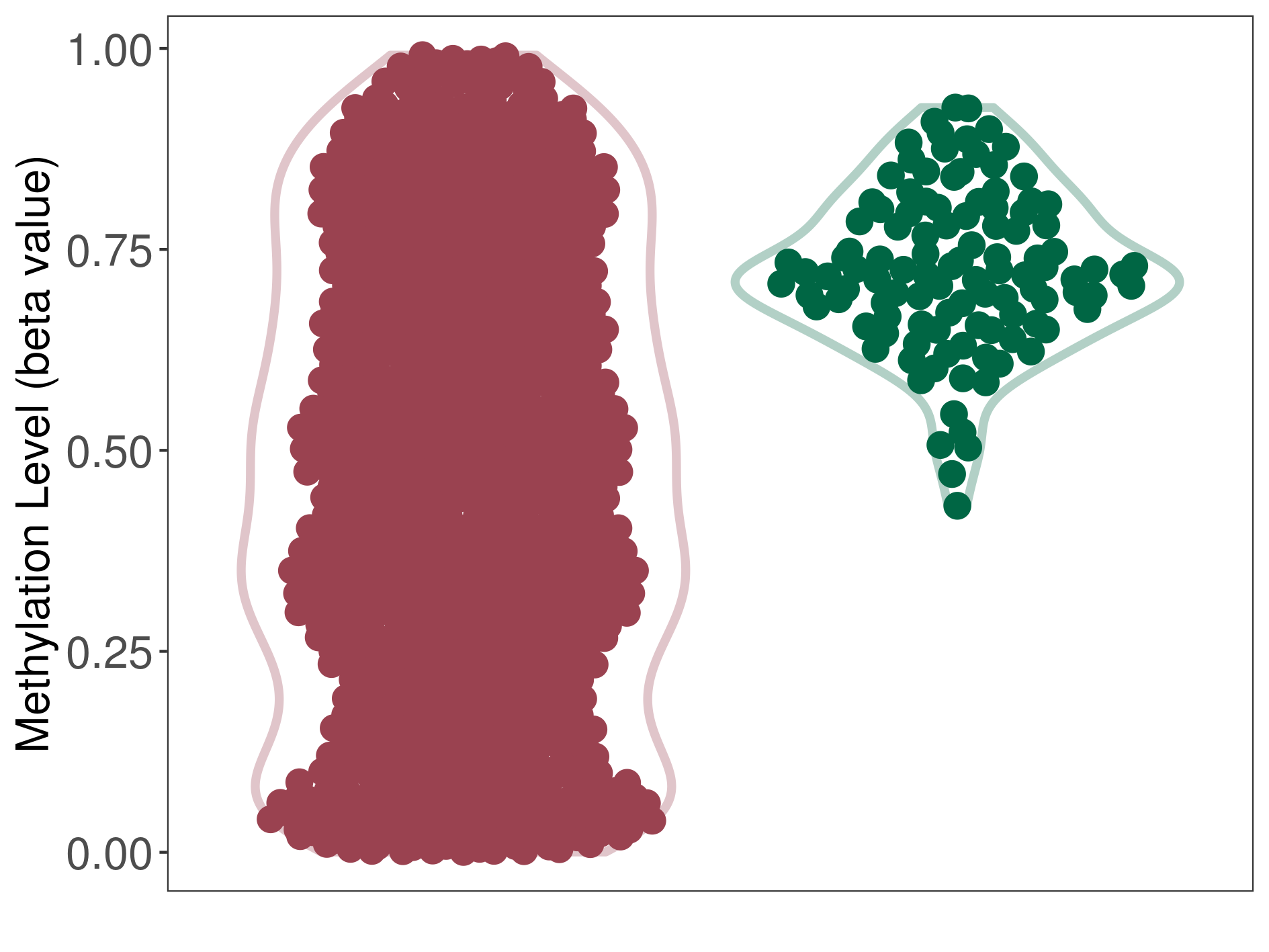
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Brain neuroblastoma | Significant hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 1.50E-07; delta-beta: -3.34E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the CYP2E1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
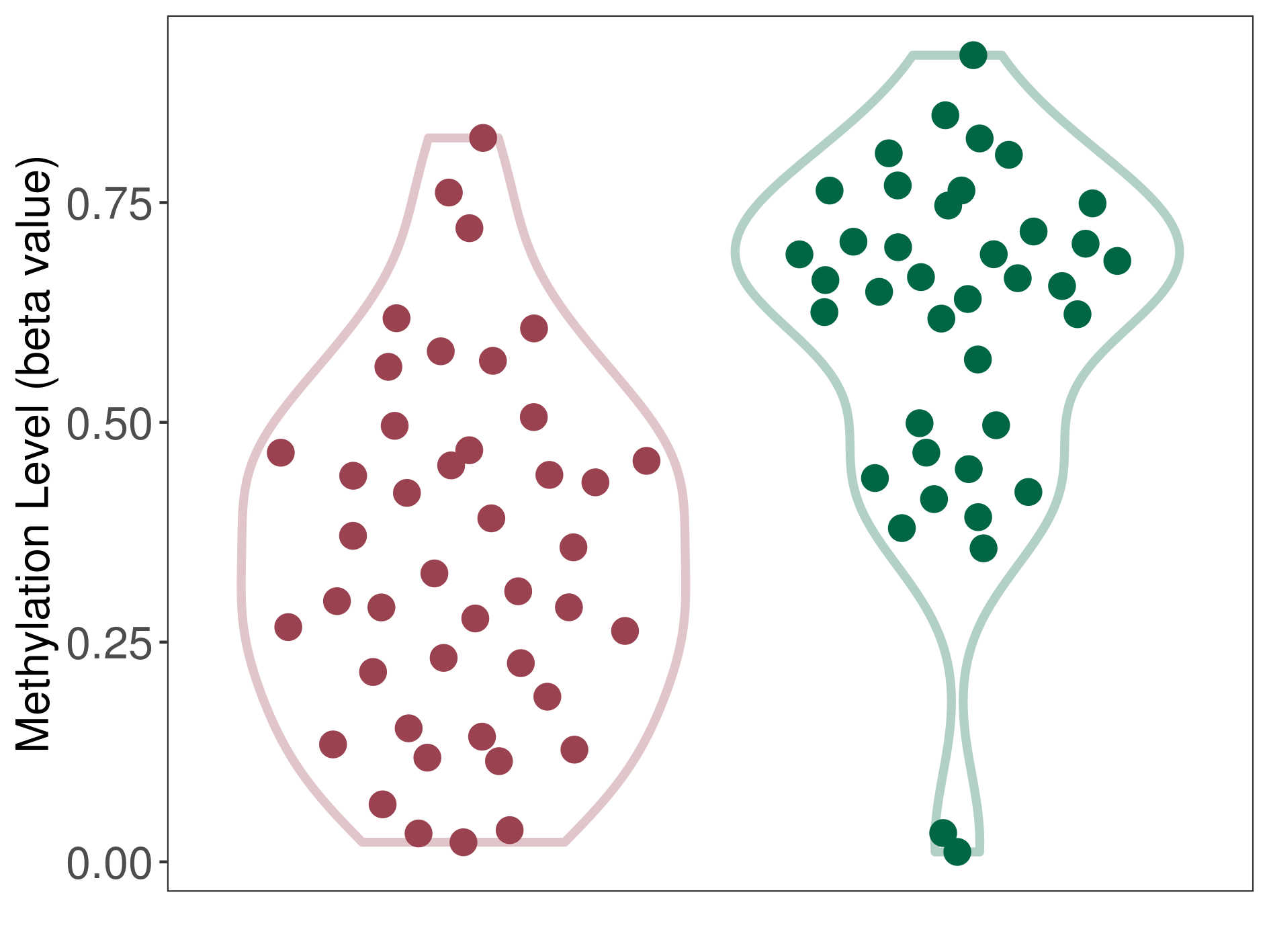
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Central neurocytoma | Moderate hypermethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypermethylation p-value: 2.84E-02; delta-beta: 2.54E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hyper-methylate the CYP2E1 gene, which leads to a moderatly decreased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
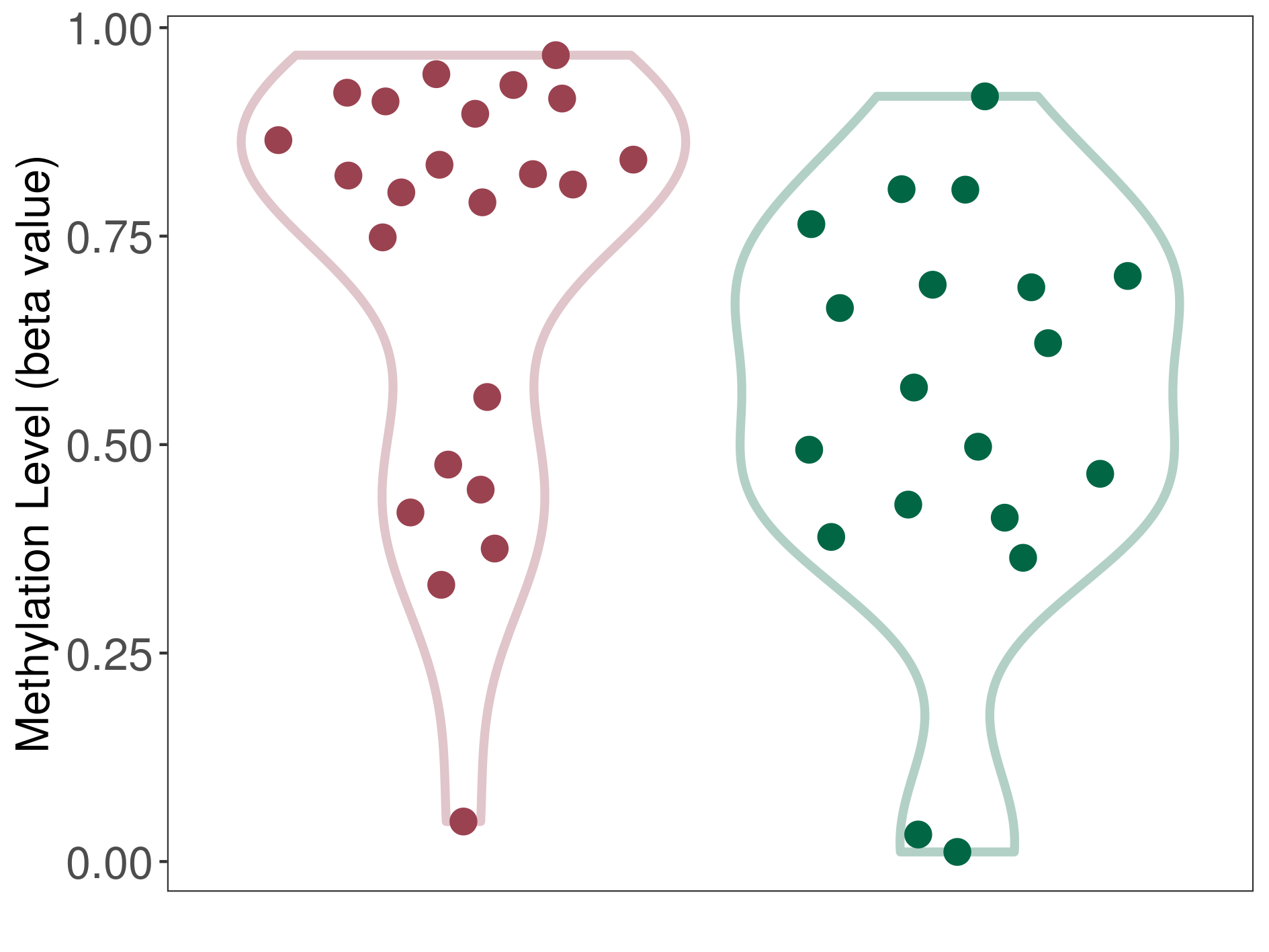
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Multilayered rosettes embryonal tumour | Significant hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 8.50E-21; delta-beta: -6.09E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the CYP2E1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
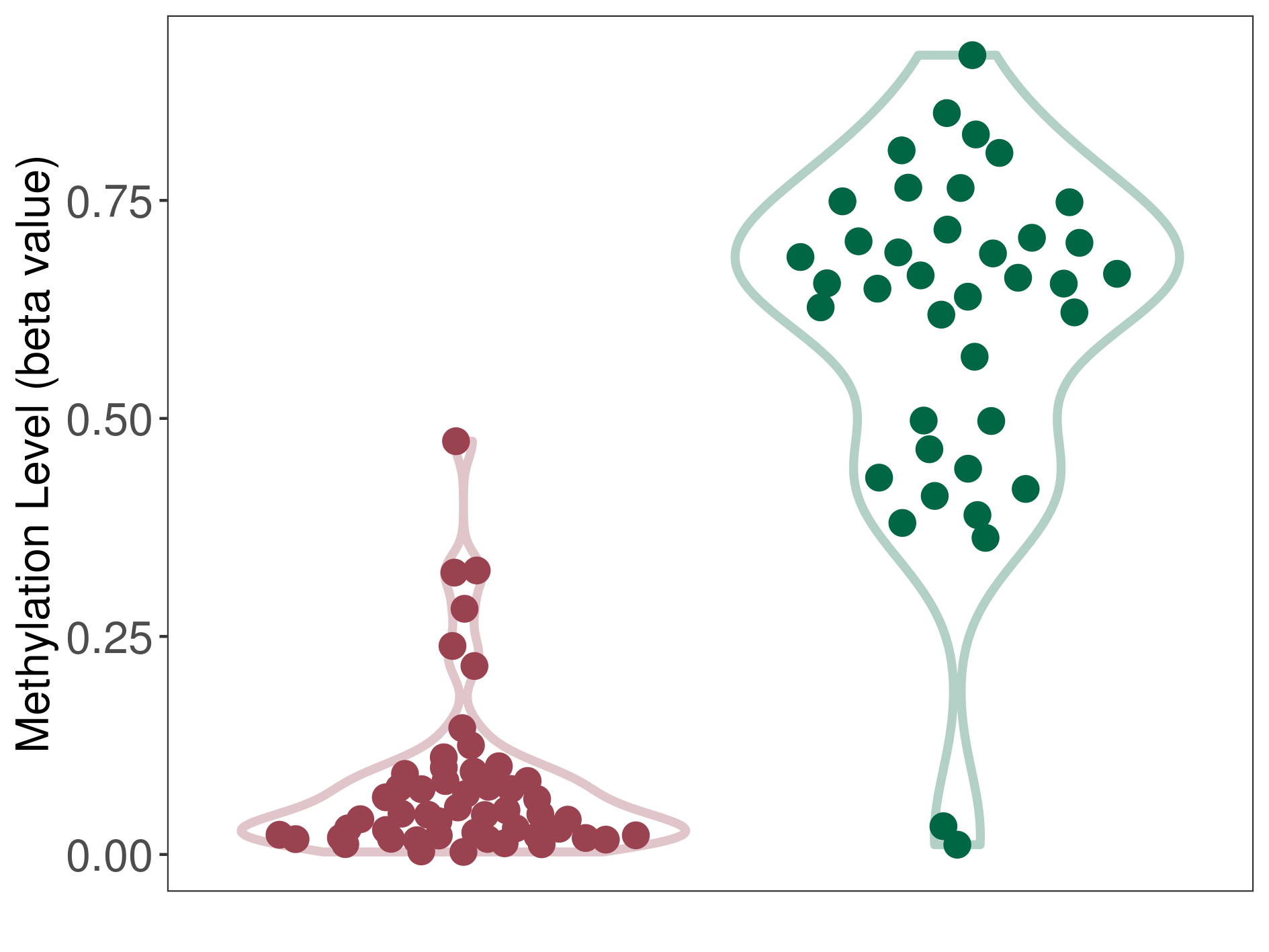
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Cerebellar liponeurocytoma | Significant hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 2.33E-04; delta-beta: -5.23E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the CYP2E1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
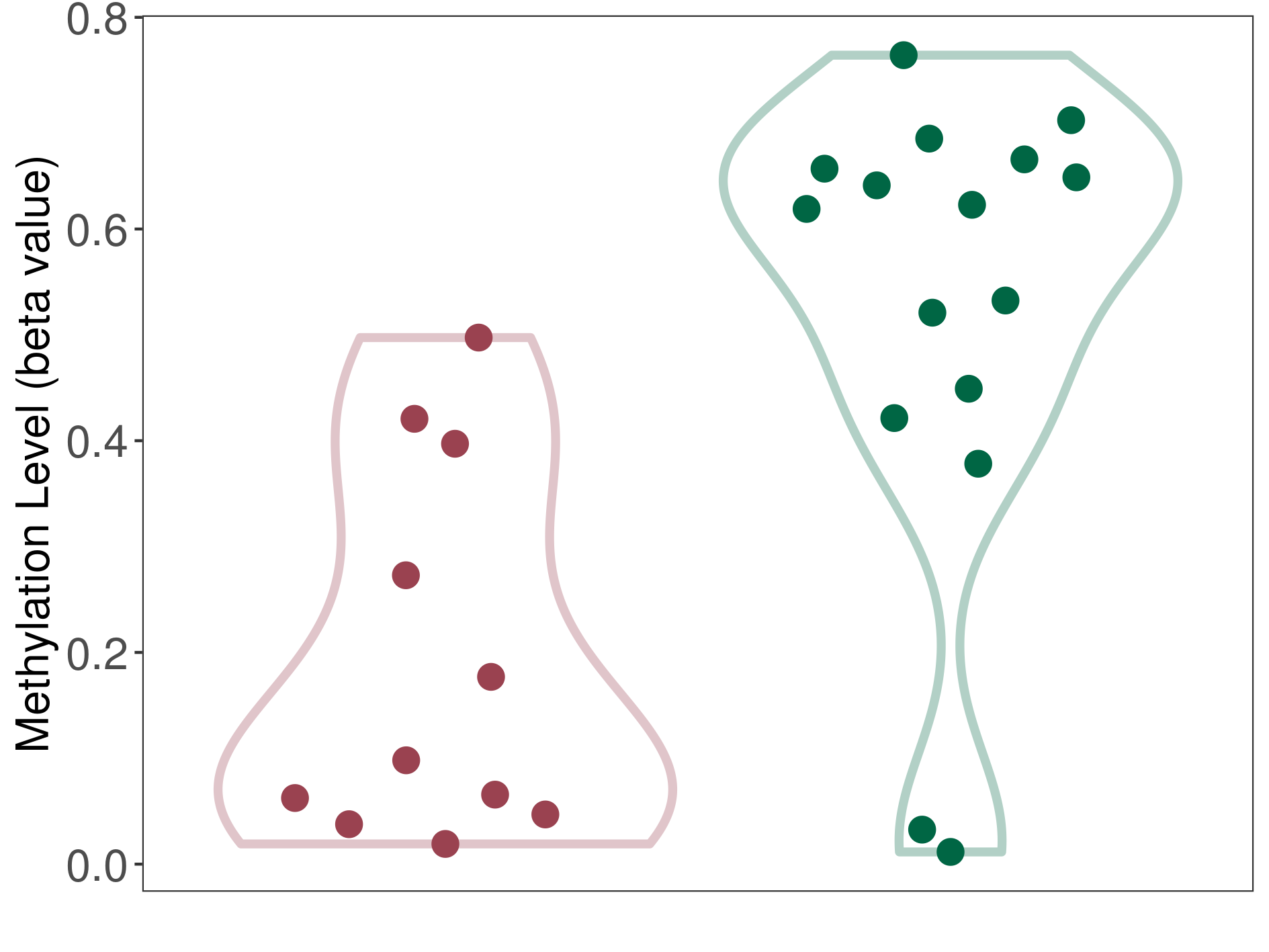
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Diffuse midline glioma | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 8.44E-13; delta-beta: -2.18E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP2E1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
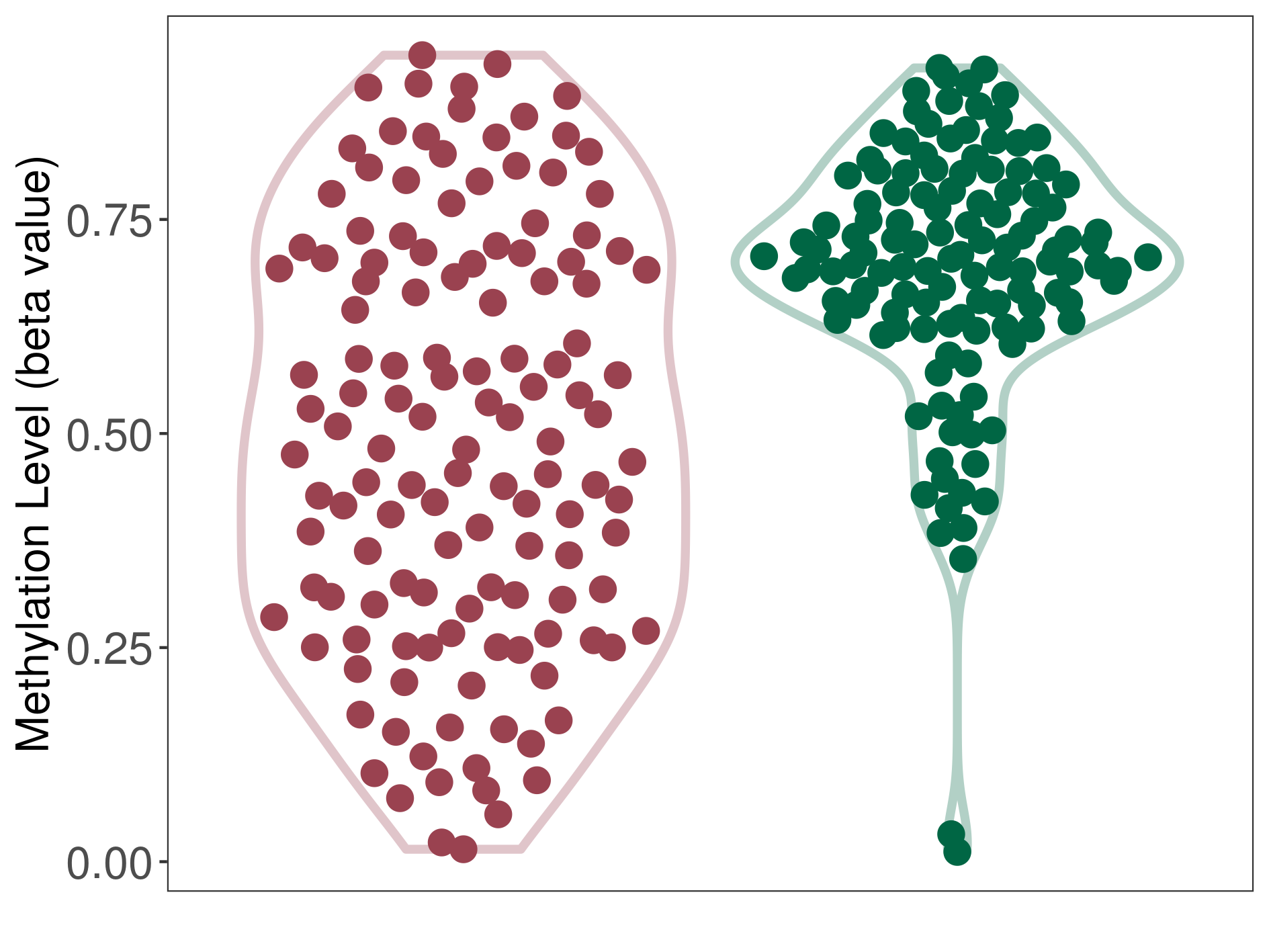
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Third ventricle chordoid glioma | Significant hypermethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypermethylation p-value: 6.00E-03; delta-beta: 4.15E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hyper-methylate the CYP2E1 gene, which leads to a significantly decreased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
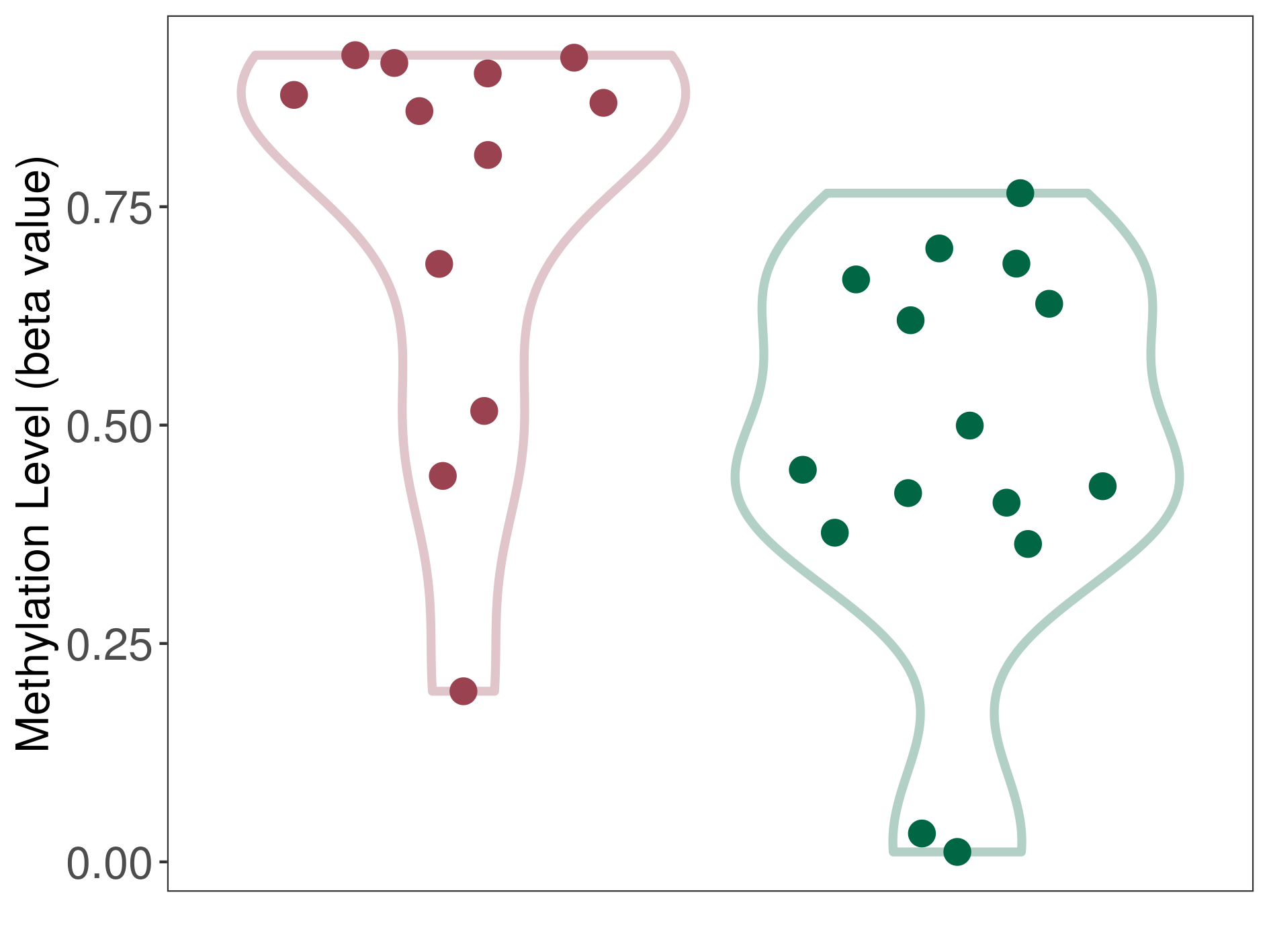
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Oligodendroglioma | Moderate hypermethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypermethylation p-value: 7.72E-14; delta-beta: 2.12E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hyper-methylate the CYP2E1 gene, which leads to a moderatly decreased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
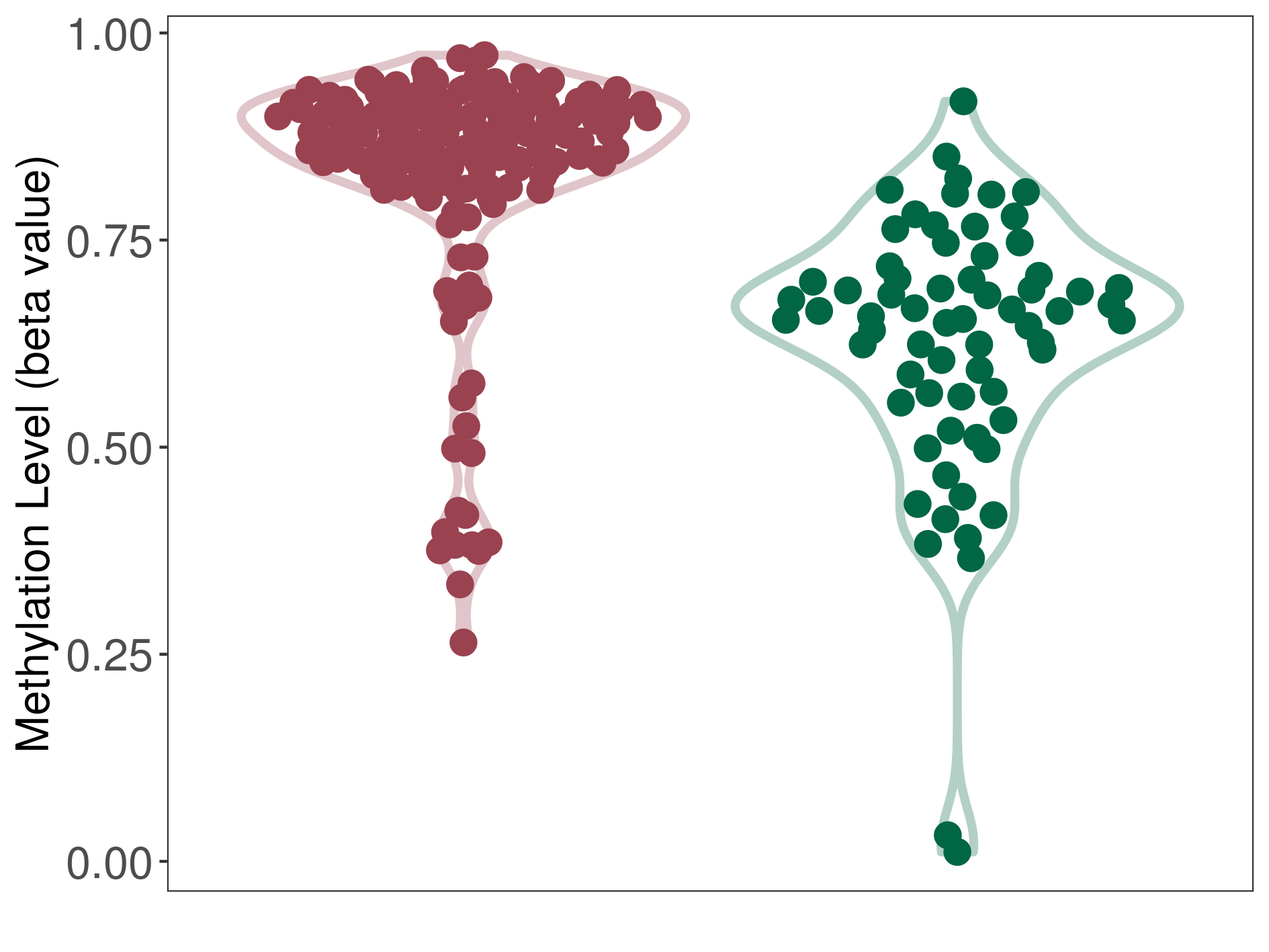
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2B5F Uterine carcinosarcoma | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Uterine carcinosarcoma | Significant hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 5.11E-04; delta-beta: -3.36E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the CYP2E1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
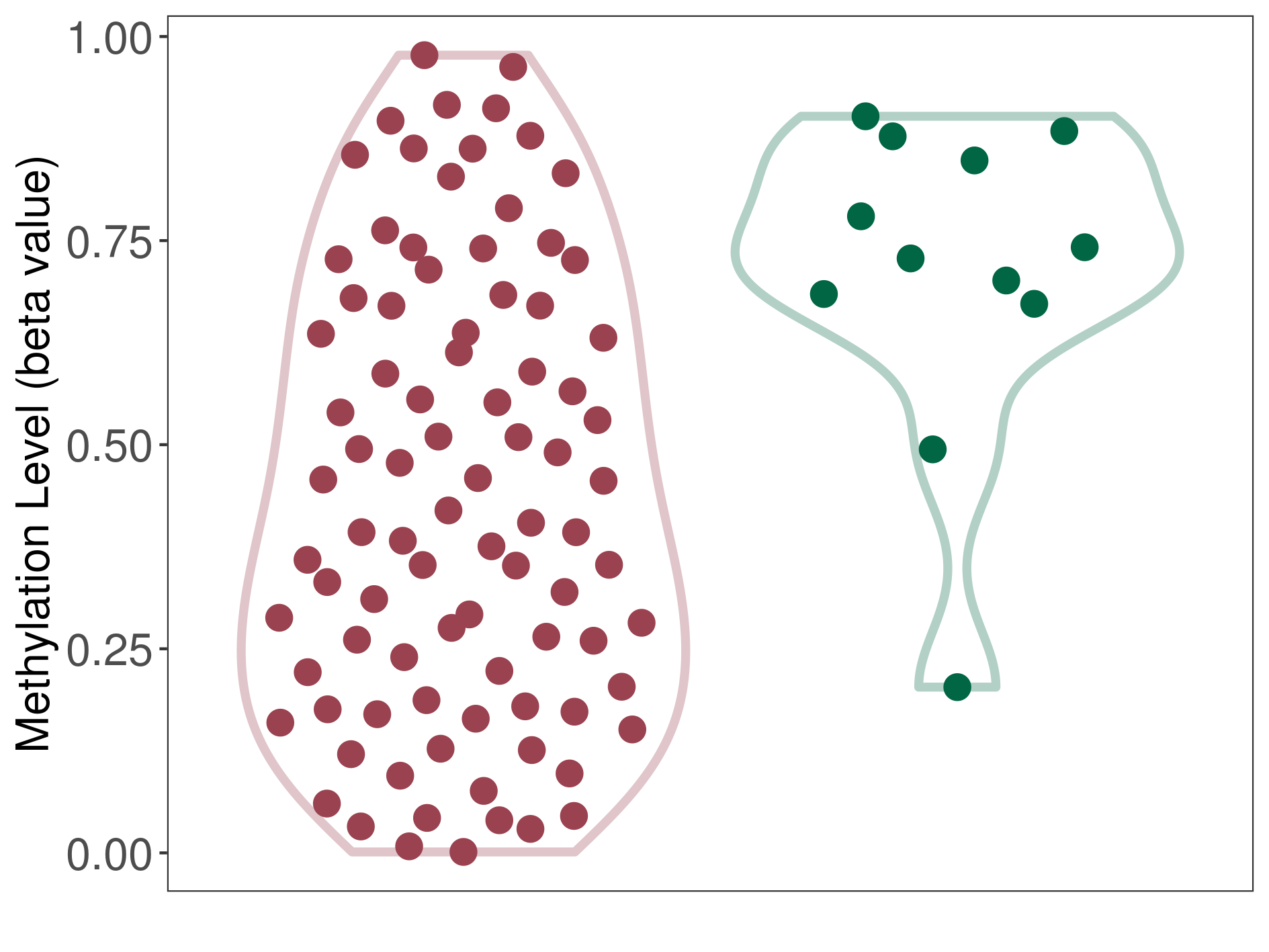
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2B60 Squamous cell carcinoma | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Squamous cell carcinoma | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 4.98E-04; delta-beta: -2.65E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP2E1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue adjacent to the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
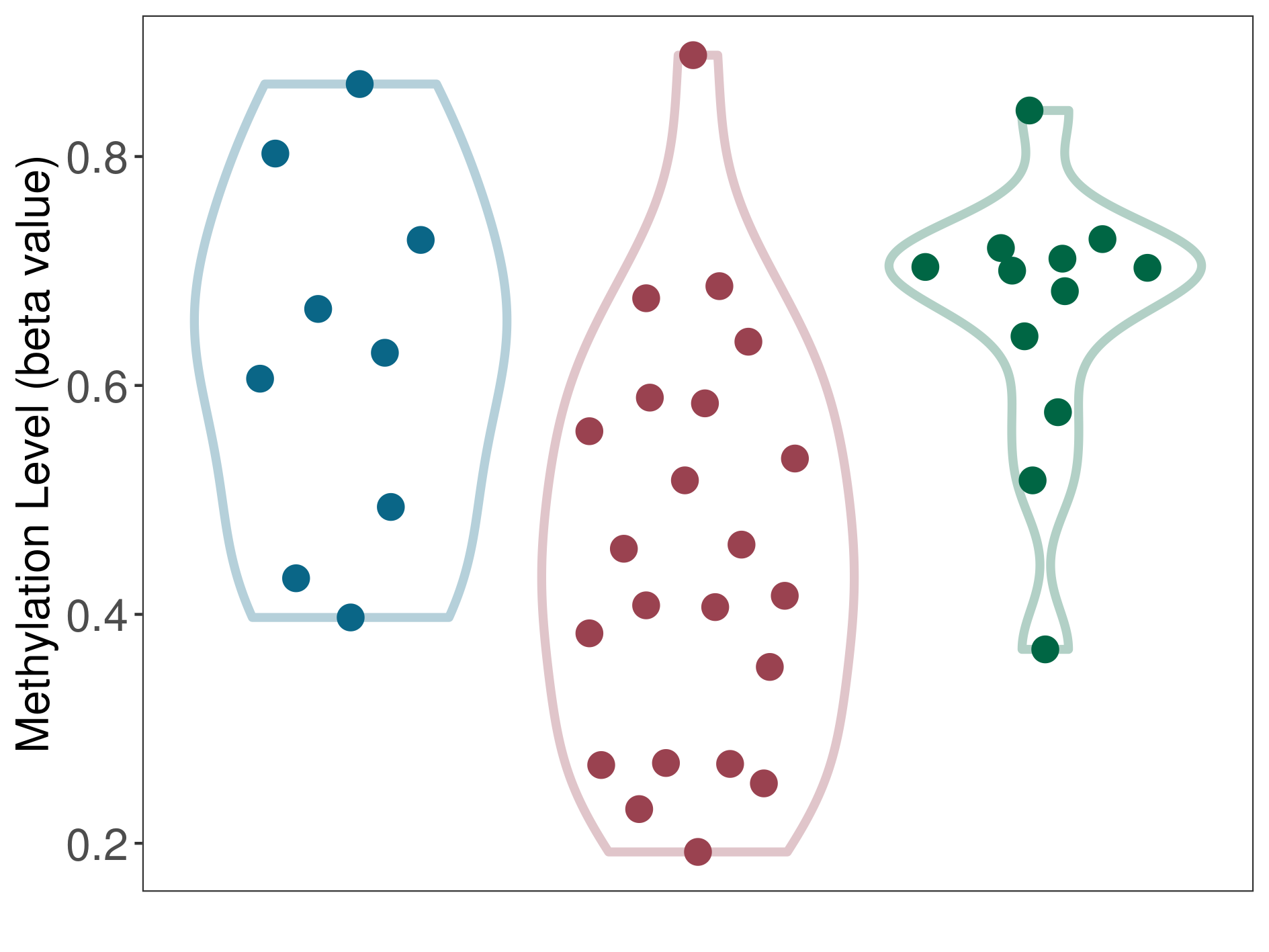
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C12 Liver cancer | Click to Show/Hide the Full List of HOSPPI: 5 HOSPPI | ||||
| Oligomerization | |||||
| Progesterone receptor 1 (PGRMC1) | Liver cancer | Heterooligomer | |||
| Uniprot ID | |||||
| Interaction Name | PGRMC1-CYP2E1 heterooligomerization | [10] | |||
| Studied Cell Lines | Human liver cancer cell line (HepG2) | ||||
| Affected Substrate(s): | Chlorzoxazone (Metabolic product: Chlorzoxazone 6-hydroxylase) 7-ethoxycoumarin (Metabolic product: 7-ethoxycoumarin O-deethylase) | ||||
| Description | Progesterone receptor 1 (PGRMC1) is reported to heterooligomerize with the CYP2E1 protein, which leads to a slight effect on the enzyme activity of Cytochrome P450 2E1. As a result, the interaction between PGRMC1 and CYP2E1 can slightly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Transcription-factor regulation | |||||
| NF of activated T-cell 1 (NFATC1) | Liver cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | NFATC1-CYP2E1 interaction | [9] | |||
| Studied Cell Lines | B16A2 cell line | ||||
| Ensembl ID | |||||
| Description | NF of activated T-cell 1 (NFATC1) is reported to activate the transcription of CYP2E1 gene, which leads to an increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between NFATC1 and CYP2E1 can activate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| STAT 6 (STAT6) | Liver cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | STAT6-CYP2E1 interaction | [9] | |||
| Studied Cell Lines | B16A2 cell line | ||||
| Ensembl ID | |||||
| Description | STAT 6 (STAT6) is reported to activate the transcription of CYP2E1 gene, which leads to an increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between STAT6 and CYP2E1 can activate the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| Histone modification | |||||
| Histone acetyltransferases (HATs) | Liver cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | HATs-CYP2E1 interaction | [8] | |||
| Studied Cell Lines | HepG2 cell line | ||||
| Description | Histone acetyltransferases (HATs) are reported to acetylate the CYP2E1 gene and thereby activate the transcriptional activity of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between HATs and CYP2E1 can enhance the drug-metabolizing process of Cytochrome P450 2E1. | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Liver cancer | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Adjacent Tissue | Moderate hypomethylation p-value: 2.85E-07; delta-beta: -2.02E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP2E1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue adjacent to the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
DME methylation in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
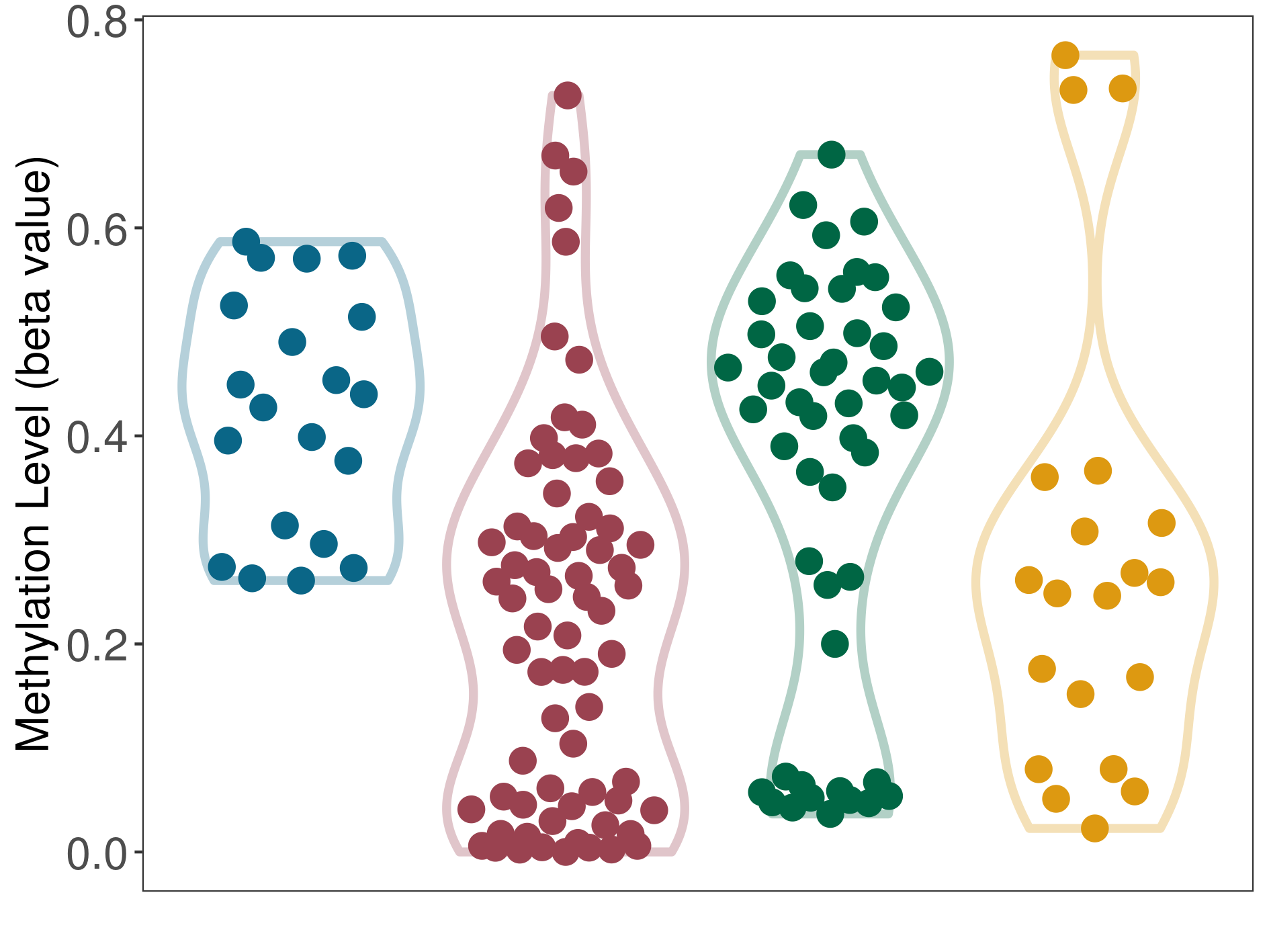
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C30 Melanoma | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Melanoma | Significant hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 1.64E-03; delta-beta: -3.28E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the CYP2E1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
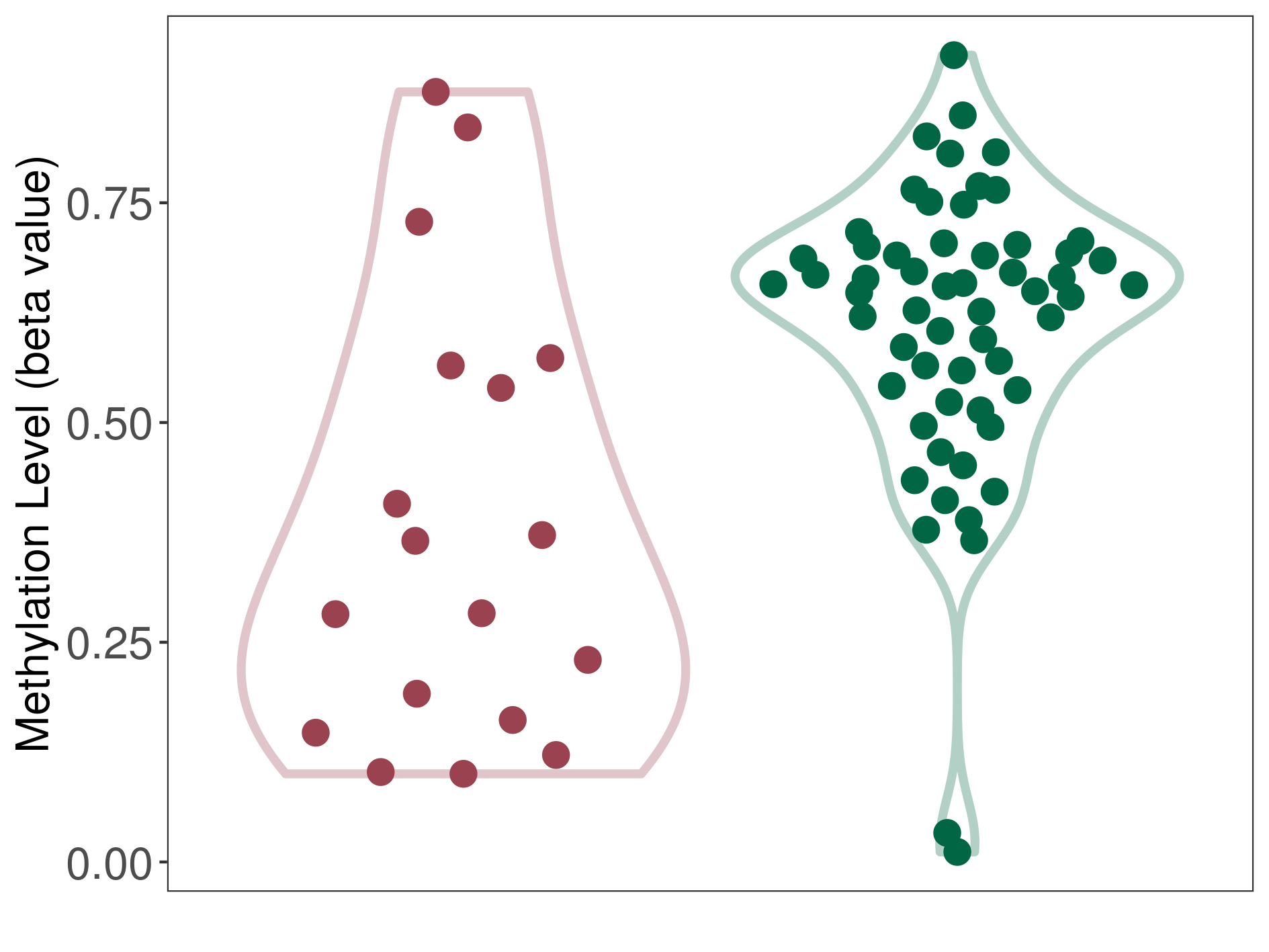
|
Click to View the Clearer Original Diagram | |||
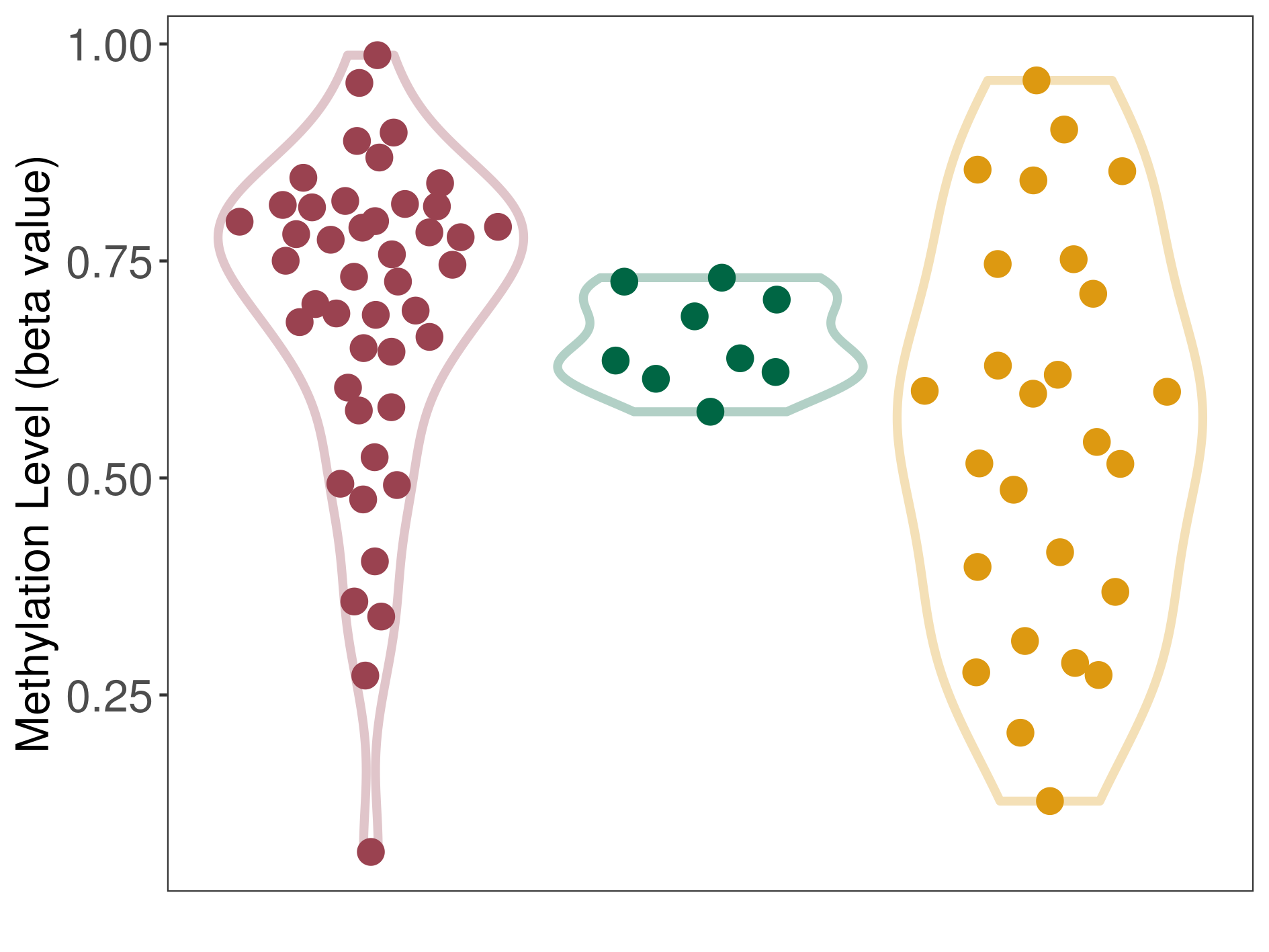
| ICD-11: 2C90 Renal cell carcinoma | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Renal cell carcinoma | Significant hypermethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Other Disease Section | Significant hypermethylation p-value: 1.55E-02; delta-beta: 1.76E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hyper-methylate the CYP2E1 gene, which leads to a significantly decreased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can significantly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
DME methylation in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 05 Endocrine/nutritional/metabolic diseases | |||||
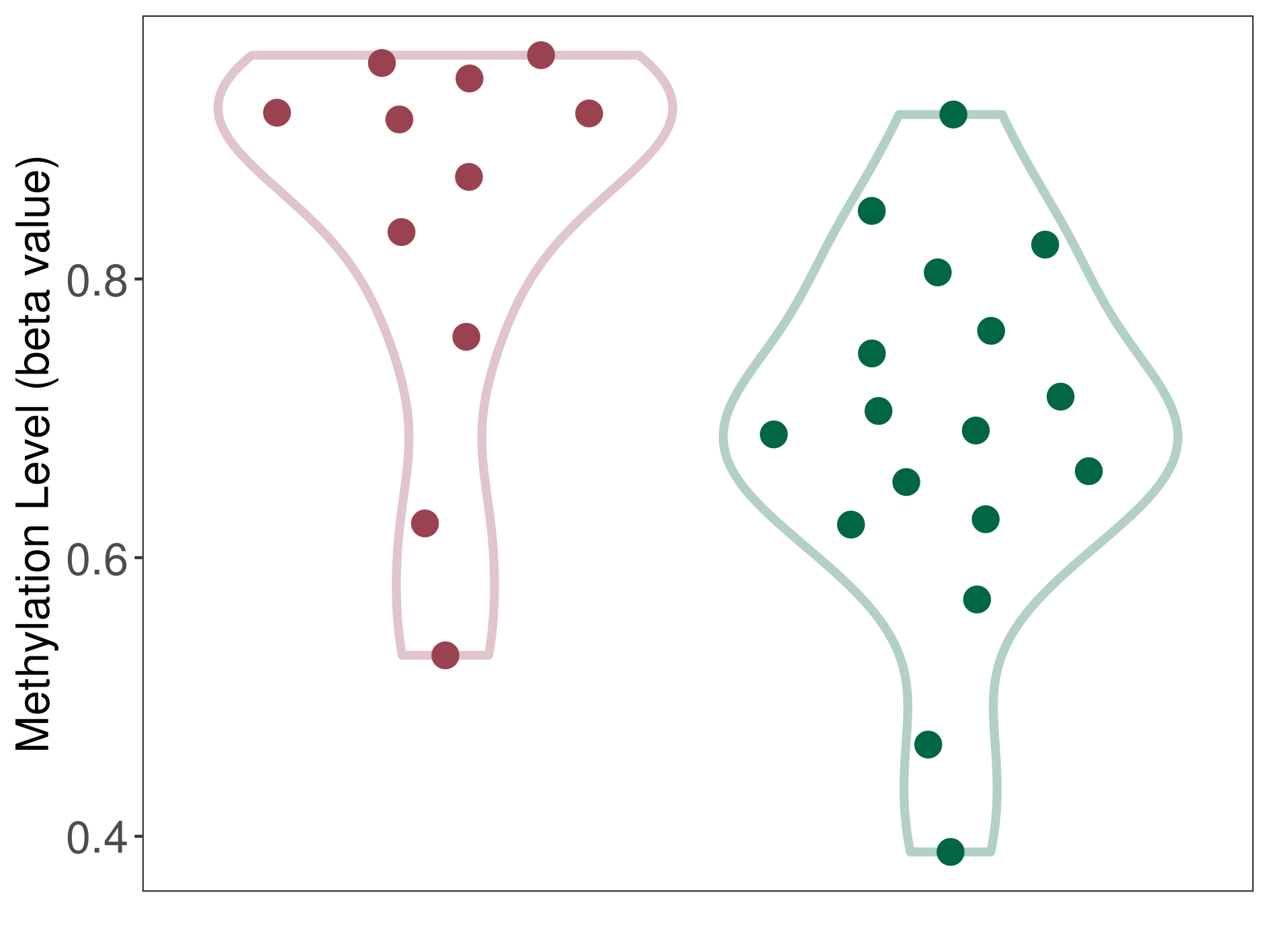
| ICD-11: 5A61 Pituitary gland hypofunction | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Chordoma | Moderate hypermethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypermethylation p-value: 1.11E-02; delta-beta: 2.23E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hyper-methylate the CYP2E1 gene, which leads to a moderatly decreased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
|
Click to View the Clearer Original Diagram | |||
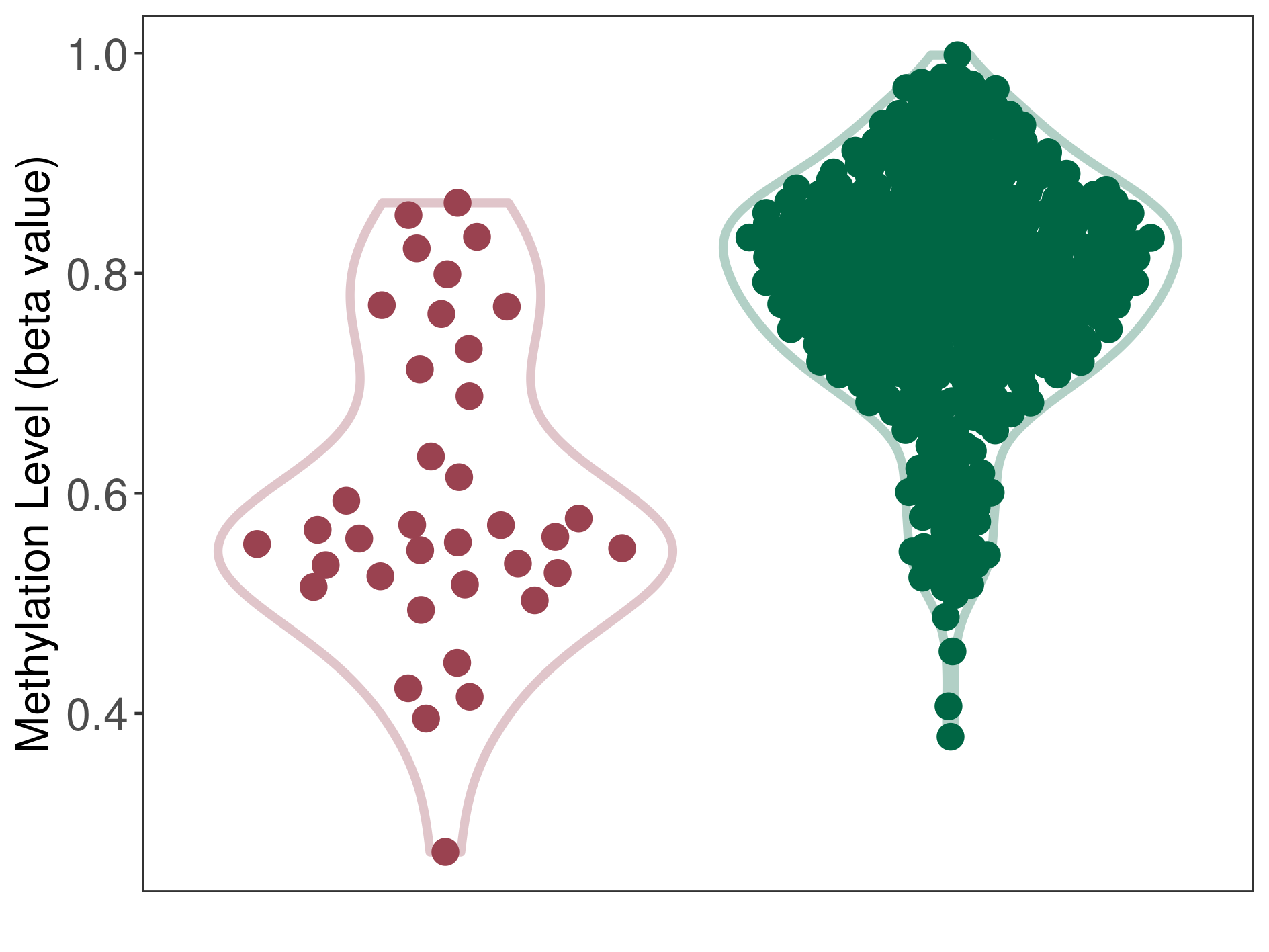
| ICD-11: 5B81 Obesity | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Obesity | Moderate hypomethylation | |||
| Interaction Name | DNMT-CYP2E1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Moderate hypomethylation p-value: 1.72E-09; delta-beta: -2.35E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to moderatly hypo-methylate the CYP2E1 gene, which leads to a moderatly increased expression of the drug-metabolizing enzyme Cytochrome P450 2E1. As a result, the interaction between DNMT and CYP2E1 can moderatly affect the drug-metabolizing process of Cytochrome P450 2E1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
|
Click to View the Clearer Original Diagram | |||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

