Details of Host Protein-DME Interaction (HOSPPI)
| General Information of Drug-Metabolizing Enzyme (DME ID: DME0461) | |||||
|---|---|---|---|---|---|
| DME Name | Prolyl 4-hydroxylase alpha-1 (P4HA1), Homo sapiens | DME Info | |||
| UniProt ID | |||||
| EC Number | EC: 1.14.11.2 (Click to Show/Hide the Complete EC Tree) | ||||
| Lineage | Species: Homo sapiens (Click to Show/Hide the Complete Species Lineage) | ||||
| Interactome | |||||
| Disease Specific Interactions between Host Protein and DME (HOSPPI) | |||||
|---|---|---|---|---|---|
| ICD Disease Classification Healthy | |||||
| ICD-11: Healthy | Click to Show/Hide the Full List of HOSPPI: 2 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Transcription factor Sp1 (SP1) | Health | Activation | |||
| Uniprot ID | |||||
| Interaction Name | SP1-P4HA1 interaction | [2] | |||
| Studied Cell Lines | Human aortic smooth muscle cell line | ||||
| Ensembl ID | |||||
| Description | Transcription factor Sp1 (SP1) is reported to activate the transcription of P4HA1 gene, which leads to an increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between SP1 and P4HA1 can activate the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-122-5p | Health | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-122-5p--P4HA1 regulation | [1] | |||
| Studied Cell Lines | Hepatic stellate cells | ||||
| Description | hsa-miR-122-5p is reported to suppress P4HA1 mRNA translation by binding to the 3' untranslated region (3'UTR) of P4HA1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. | ||||
| ICD Disease Classification 02 Neoplasms | |||||
| ICD-11: 2A00 Brain cancer | Click to Show/Hide the Full List of HOSPPI: 4 HOSPPI | ||||
| DNA methylation | |||||
| DNA methyltransferase (DNMT) | Brain neuroblastoma | Significant hypomethylation | |||
| Interaction Name | DNMT-P4HA1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 2.48E-10; delta-beta: -3.79E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the P4HA1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between DNMT and P4HA1 can significantly affect the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
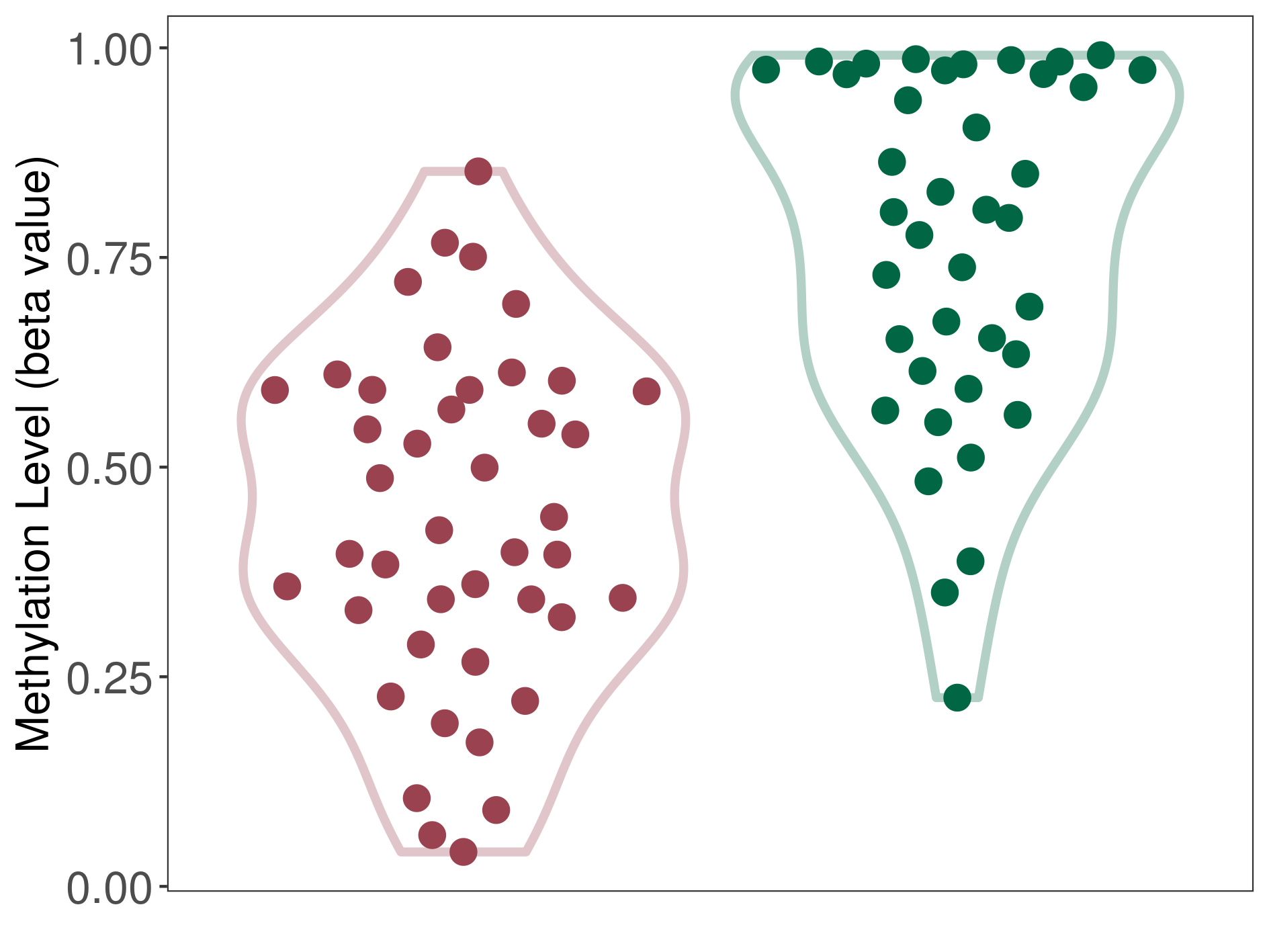
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Brain neuroepithelial tumour | Significant hypomethylation | |||
| Interaction Name | DNMT-P4HA1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 2.08E-09; delta-beta: -4.93E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the P4HA1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between DNMT and P4HA1 can significantly affect the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
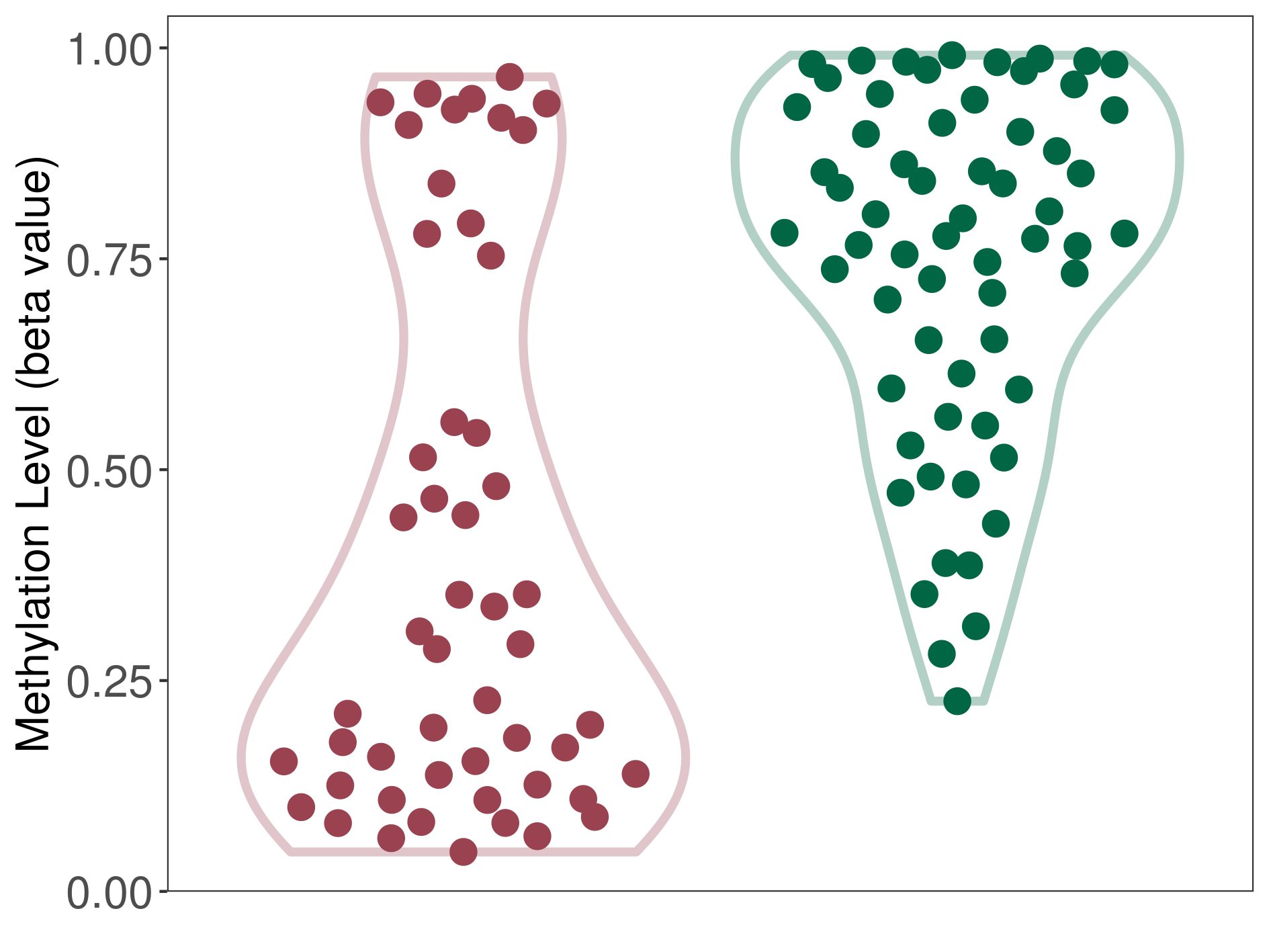
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Third ventricle chordoid glioma | Significant hypomethylation | |||
| Interaction Name | DNMT-P4HA1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 7.73E-03; delta-beta: -4.89E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the P4HA1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between DNMT and P4HA1 can significantly affect the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
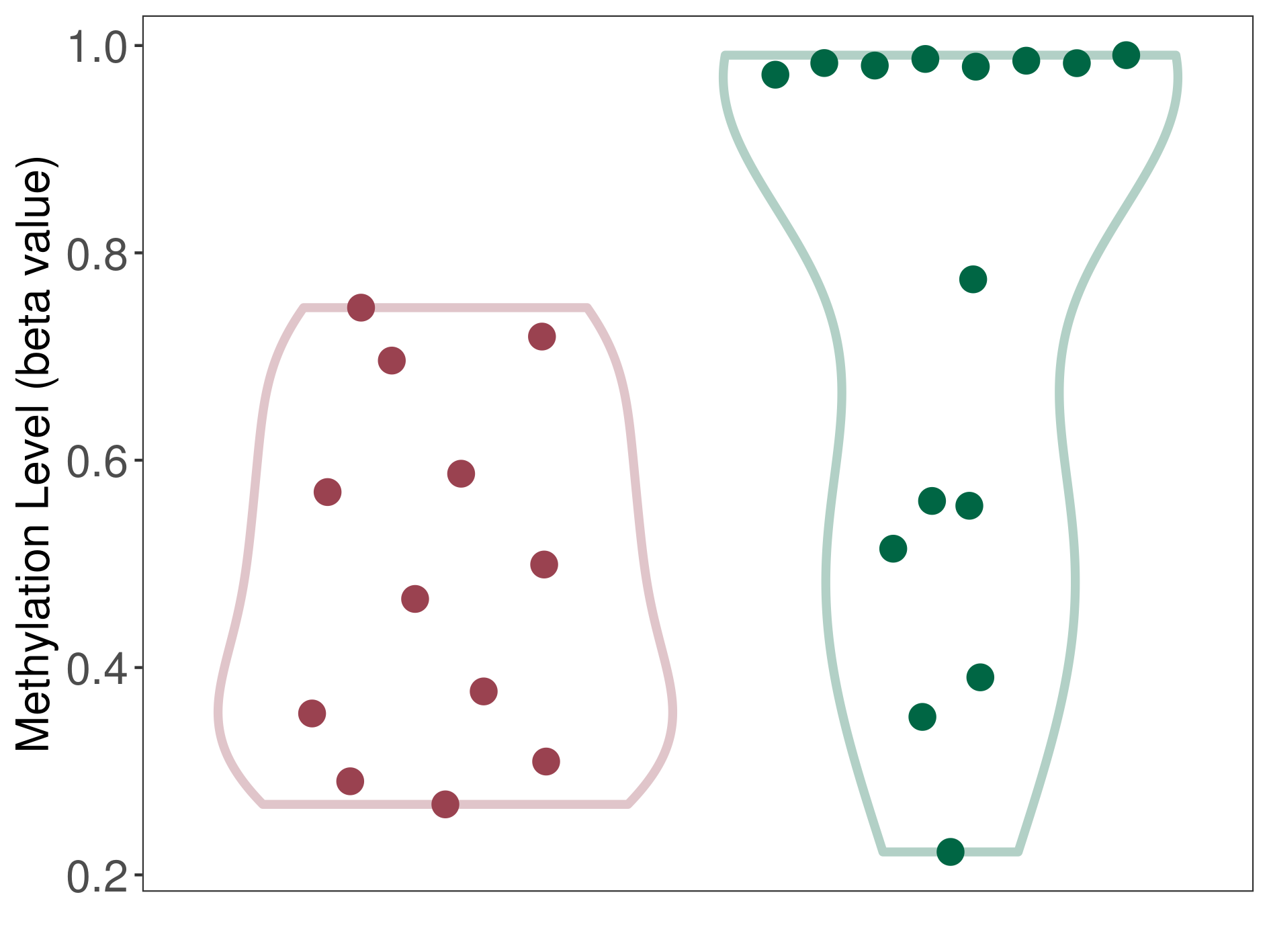
|
Click to View the Clearer Original Diagram | |||
| DNA methyltransferase (DNMT) | Spinal ependymoma | Significant hypomethylation | |||
| Interaction Name | DNMT-P4HA1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 5.97E-10; delta-beta: -3.74E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the P4HA1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between DNMT and P4HA1 can significantly affect the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
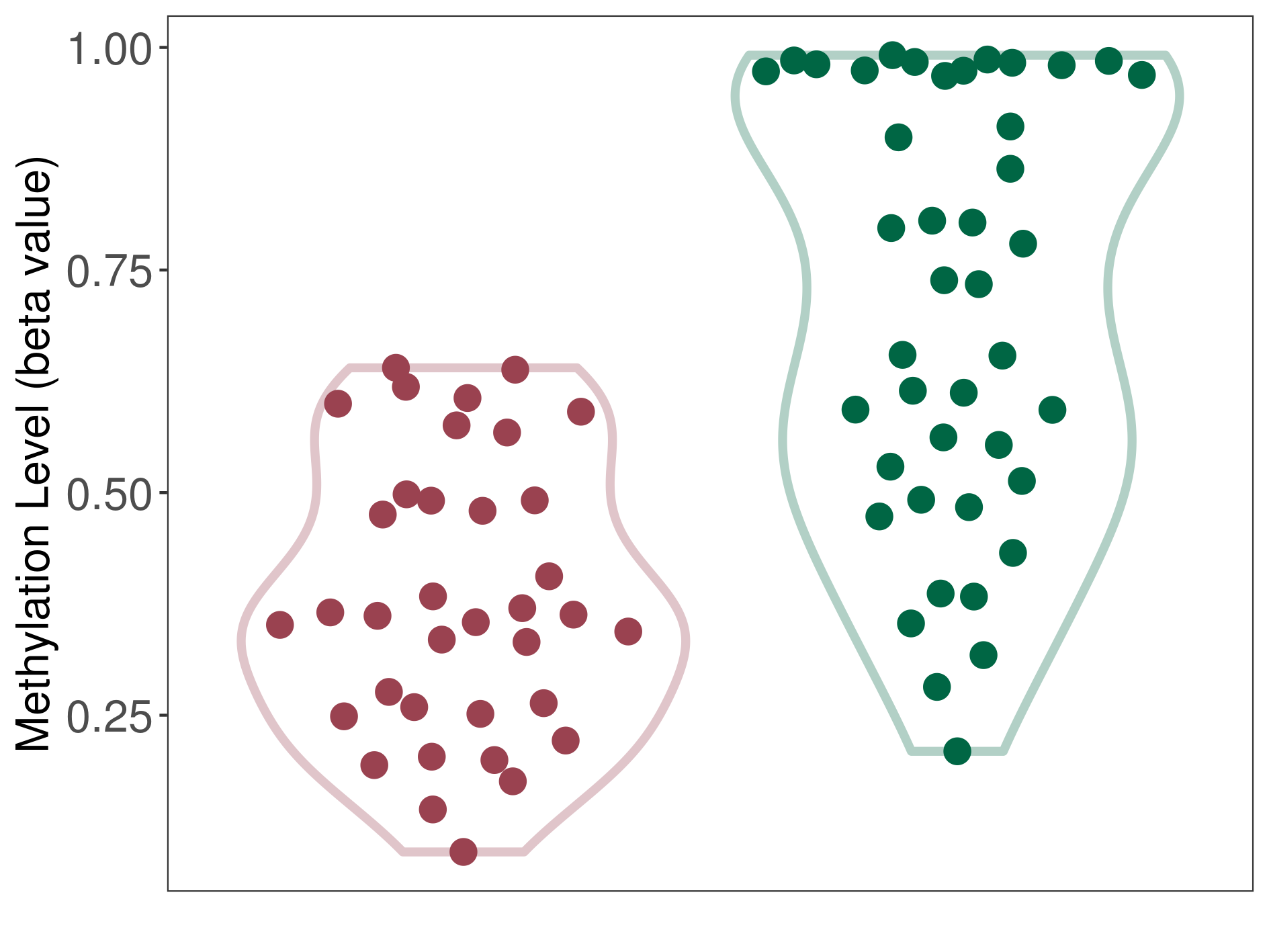
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C12 Liver cancer | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| Non-coding RNA regulation | |||||
| hsa-miR-30e-5p | Liver cancer | Suppression | |||
| miRBase ID | |||||
| Interaction Name | hsa-miR-30e-5p--P4HA1 regulation | [3] | |||
| Studied Cell Lines | HepG2 cell line | ||||
| Description | hsa-miR-30e-5p is reported to suppress P4HA1 mRNA translation by binding to the 3' untranslated region (3'UTR) of P4HA1 mRNA, which leads to a decreased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. | ||||
| ICD-11: 2C77 Cervical cancer | Click to Show/Hide the Full List of HOSPPI: 2 HOSPPI | ||||
| Transcription-factor regulation | |||||
| Upstream stimulatory 1 (USF1) | Cervical cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | USF1-P4HA1 interaction | [4] | |||
| Studied Cell Lines | HeLa and Human aortic vascular smooth muscle cell lines | ||||
| Ensembl ID | |||||
| Description | Upstream stimulatory 1 (USF1) is reported to activate the transcription of P4HA1 gene, which leads to an increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between USF1 and P4HA1 can activate the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
| Upstream stimulatory 2 (USF2) | Cervical cancer | Activation | |||
| Uniprot ID | |||||
| Interaction Name | USF2-P4HA1 interaction | [4] | |||
| Studied Cell Lines | HeLa and Human aortic vascular smooth muscle cell lines | ||||
| Ensembl ID | |||||
| Description | Upstream stimulatory 2 (USF2) is reported to activate the transcription of P4HA1 gene, which leads to an increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between USF2 and P4HA1 can activate the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
| ICD-11: 2D50 Brain cancer metastasis | Click to Show/Hide the Full List of HOSPPI: 1 HOSPPI | ||||
| DNA methylation | |||||
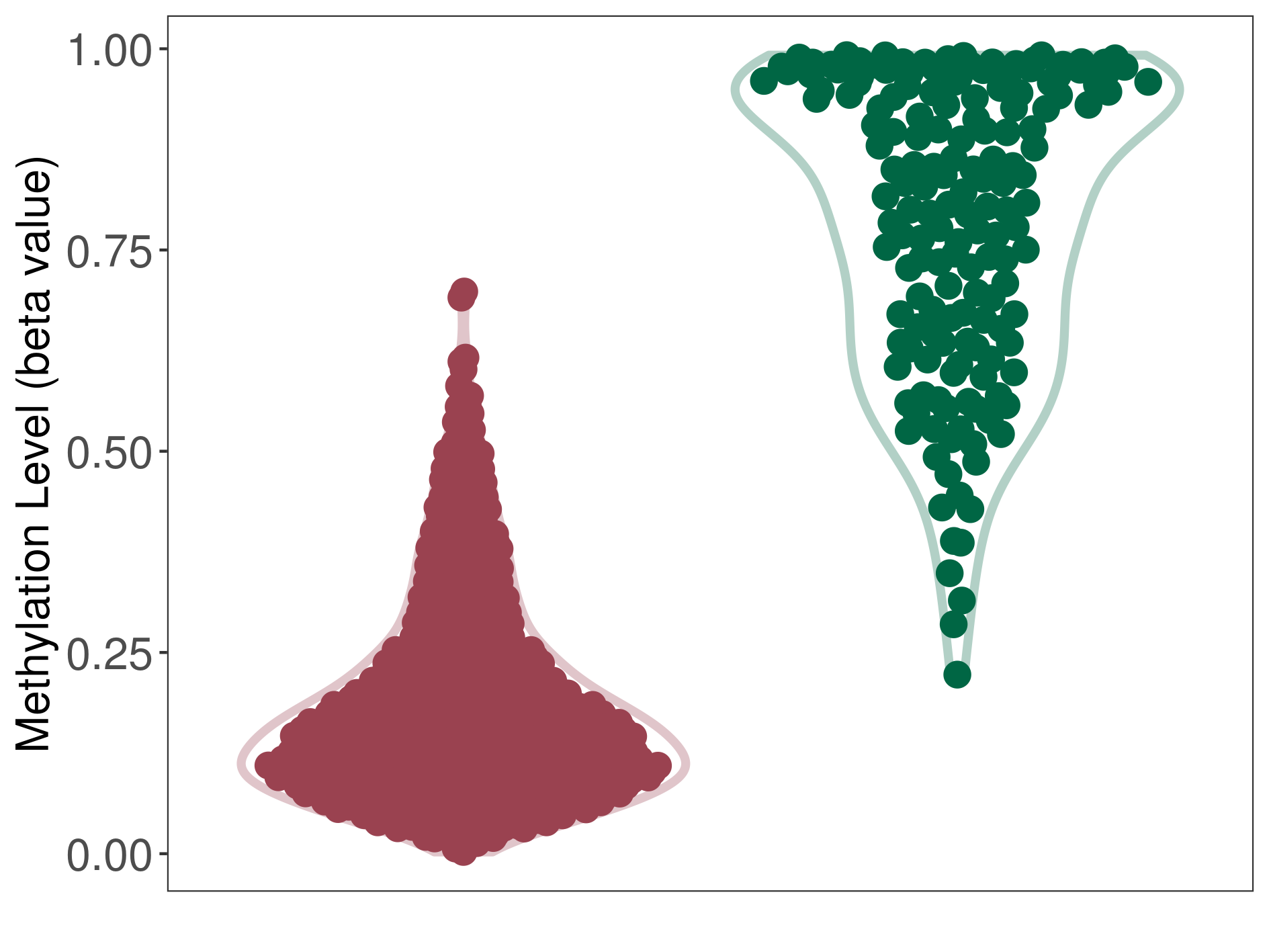
| DNA methyltransferase (DNMT) | Posterior fossa ependymoma | Significant hypomethylation | |||
| Interaction Name | DNMT-P4HA1 interaction | ||||
| The Methylation Level of Disease Section Compare with the Healthy Individual Tissue | Significant hypomethylation p-value: 1.10E-94; delta-beta: -6.76E-01 | ||||
| Description | DNA methyltransferase (DNMT) is reported to significantly hypo-methylate the P4HA1 gene, which leads to a significantly increased expression of the drug-metabolizing enzyme Prolyl 4-hydroxylase alpha-1. As a result, the interaction between DNMT and P4HA1 can significantly affect the drug-metabolizing process of Prolyl 4-hydroxylase alpha-1. | ||||
|
DME methylation in the diseased tissue of patients
DME methylation in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Methylation Level |
|
Click to View the Clearer Original Diagram | |||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

