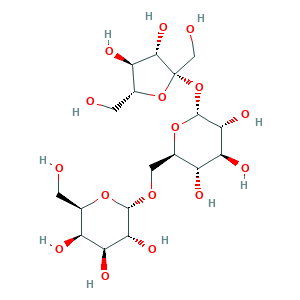
| Synonyms |
D-Raffinose; Gossypose; MUPFEKGTMRGPLJ-ZQSKZDJDSA-N; Melitose; Melitriose; N5O3QU595M; RAFFINOSE; Raffinose (8CI); Raffinose, pure; d(+)-raffinose; d-(+)-Raffinose; rafinose; raflinose; 512-69-6; 6G-alpha-D-galactosylsucrose; AI3-19427; C18H32O16; CHEBI:16634; EINECS 208-146-9; NSC 170228; NSC 2025; NSC170228; UNII-N5O3QU595M; alpha-D-glucopyranoside, beta-D-fructofuranosyl O-alpha-D-galactopyranosyl-(1->6)-; beta-D-fructofuranosyl alpha-D-galactopyranosyl-(1->6)-alpha-D-glucopyranoside
|
| Cross-matching ID |
- PubChem CID
- 439242
- ChEBI ID
-
- CAS Number
-
- Formula
- C18H32O16
- Canonical SMILES
- C(C1C(C(C(C(O1)OCC2C(C(C(C(O2)OC3(C(C(C(O3)CO)O)O)CO)O)O)O)O)O)O)O
- InChI
- 1S/C18H32O16/c19-1-5-8(22)11(25)13(27)16(31-5)30-3-7-9(23)12(26)14(28)17(32-7)34-18(4-21)15(29)10(24)6(2-20)33-18/h5-17,19-29H,1-4H2/t5-,6-,7-,8+,9-,10-,11+,12+,13-,14-,15+,16+,17-,18+/m1/s1
- InChIKey
- MUPFEKGTMRGPLJ-ZQSKZDJDSA-N
|