| Synonyms |
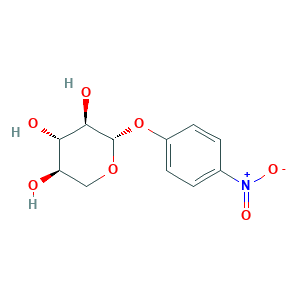
P-nitrophenyl-beta-D-xyloside; Nitrophenyl-beta-D-xyloside; p-Nitrophenyl beta-D-xyloside; p-nitrophenyl ss-D-xylopyranoside; (2S,3R,4S,5R)-2-(4-Nitrophenoxy)tetrahydro-2H-pyran-3,4,5-triol; (2S,3R,4S,5R)-2-(4-nitrophenoxy)oxane-3,4,5-triol; 2001-96-9; 3-Nitrophenyl b-D-xylopyranoside; 4-Nitrophenyl b-D-xylopyranoside; 4-Nitrophenyl beta-D-xylopyranoside; 4-Nitrophenyl beta-D-xyloside; 4-Nitrophenyl-Beta-D-xylopyranoside; 41721-77-1; NSC 371094; PNPX; p-Nitrophenyl beta-D-xylopyranoside; p-Nitrophenyl-beta-d-xylopyranoside
|
| Cross-matching ID |
- PubChem CID
- 91509
- ChEBI ID
-
- CAS Number
-
- Formula
- C11H13NO7
- Canonical SMILES
- C1C(C(C(C(O1)OC2=CC=C(C=C2)[N+](=O)[O-])O)O)O
- InChI
- 1S/C11H13NO7/c13-8-5-18-11(10(15)9(8)14)19-7-3-1-6(2-4-7)12(16)17/h1-4,8-11,13-15H,5H2/t8-,9+,10-,11+/m1/s1
- InChIKey
- MLJYKRYCCUGBBV-YTWAJWBKSA-N
|