| General Information of Drug (ID:
DR5074) |
| Drug Name |
Mitomycin
|
| Synonyms |
Mitomycin C; mitomycin C; 1950/7/7; Mutamycin; Ametycine; Mitocin-C; Ametycin; Mitomycin-C; Mytozytrex; Mitomycinum; Mytomycin; Mitozytrex; Mitomycinum C; Mitocin C; Mitomycins; Mitamycin; MMC; Mitosol; Mitomycyna C; 7-Amino-9alpha-methoxymitosane; NSC-26980; Mitomycyna C [Polish]; Mito-C; Mit-C; Mitomycin (TN); Mitomycinum [INN-Latin]; Mitomycine [INN-French]; Mitomicina [INN-Spanish]; NCI-C04706; RCRA waste number U010; NSC26980; NSC 26980; Mitomycine; CCRIS 414; UNII-50SG953SK6; HSDB 3239; C15H18N4O5; EINECS 200-008-6; Mitomycin C,; Ametycin; Mitomicina; Muamycin; Mitomycin C from Streptomyces caespitosus; Mitomycin C (JP15); Mitomycin C, Streptomyces caespitosus; Muamycin (TN); Mitomycin (USP/INN); Mitomycin [USAN:INN:BAN]; Mitomycin C, Streptomyces caespitosus, Carrier-Free
|
| Indication |
Breast cancer
[ICD11: 2C60-2C6Y]
|
Approved
|
[1]
|
|
Gastrointestinal cancer
[ICD11:
ICD11: 2C11]
|
Approved
|
[2]
|
|
Solid tumour/cancer
[ICD11:
ICD11: 2A00-2F9Z]
|
Approved
|
[3]
|
| Structure |
|
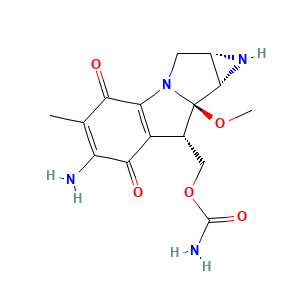 |
|
3D MOL
|
2D MOL
|
| Pharmaceutical Properties |
Molecular Weight |
334.33 |
Topological Polar Surface Area |
147 |
| Heavy Atom Count |
24 |
Rotatable Bond Count |
4 |
| Hydrogen Bond Donor Count |
3 |
Hydrogen Bond Acceptor Count |
8 |
| Cross-matching ID |
- PubChem CID
- 5746
- ChEBI ID
-
- CAS Number
-
- TTD Drug ID
- D0Y0GH
- Formula
- C15H18N4O5
- Canonical SMILES
- CC1=C(C(=O)C2=C(C1=O)N3C[C@H]4[C@@H]([C@@]3([C@@H]2COC(=O)N)OC)N4)N
- InChI
- InChI=1S/C15H18N4O5/c1-5-9(16)12(21)8-6(4-24-14(17)22)15(23-2)13-7(18-13)3-19(15)10(8)11(5)20/h6-7,13,18H,3-4,16H2,1-2H3,(H2,17,22)/t6-,7+,13+,15-/m1/s1
- InChIKey
- NWIBSHFKIJFRCO-WUDYKRTCSA-N
|
|
|
|
|
|
|
|
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.


