Details of the Drug Metabolized by Drug-Metabolizing Enzyme (DME)
| General Information of Drug (ID: DR5109) | ||||||
|---|---|---|---|---|---|---|
| Drug Name |
Oxycodone
|
|||||
| Synonyms | . | |||||
| Indication | Pain [ICD11: MG30-MG3Z] | Approved | [1] | |||
| Back pain [ICD11: ICD11: ME84] | Phase 3 | [2] | ||||
| Structure |
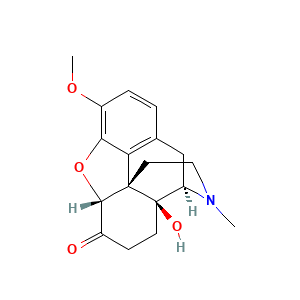 |
|||||
| 3D MOL | 2D MOL | |||||
| Pharmaceutical Properties | Molecular Weight | 315.4 | Topological Polar Surface Area | 59 | ||
| Heavy Atom Count | 23 | Rotatable Bond Count | 1 | |||
| Hydrogen Bond Donor Count | 1 | Hydrogen Bond Acceptor Count | 5 | |||
| Cross-matching ID |
|
|||||
| The Metabolic Roadmap of This Drug | |||||
|---|---|---|---|---|---|
| The Full List of Drug Metabolites (DM) of This Drug | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
| The Full List of Metabolic Reaction (MR) of This Drug | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Drug-Metabolizing Enzyme(s) (DME) Metabolizing This Drug | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

