Detail Information of Xenobiotics
| General Information of Xenobiotics (ID: XEO00079) | ||||||
|---|---|---|---|---|---|---|
| Xenobiotics Name |
Vorinostat
|
|||||
| Xenobiotics Type |
Pharmaceutical Agent(s)
|
|||||
| Classification |
Approved/Marketed Drug
|
|||||
| Structure |
<iframe style="width: 300px; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=5311"></iframe>
|
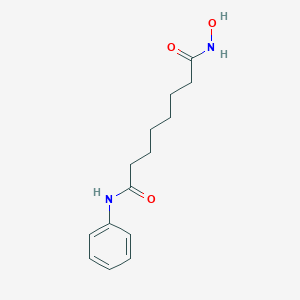 |
||||
| 3D MOL | 2D MOL | |||||
| PubChem CID | ||||||
| DME(s) Modulated by This Xenobiotics | ||||||
| DME(s) Inhibited by This Xenobiotics | ||||||
| Alcohol dehydrogenase class-I beta (ADH1B) | DME Info | Homo sapiens | [1] | |||
| Aldo-keto reductase 1C3 (AKR1C3) | DME Info | Homo sapiens | [2], [3] | |||
| Arylsulfatase B (ARSB) | DME Info | Homo sapiens | [3] | |||
| Cytochrome P450 1B1 (CYP1B1) | DME Info | Homo sapiens | [4], [5] | |||
| Retinoic acid 4-hydroxylase 26A1 (CYP26A1) | DME Info | Homo sapiens | [5] | |||
| DOPA decarboxylase (DDC) | DME Info | Homo sapiens | [2], [6] | |||
| Delta(24)-sterol reductase (DHCR24) | DME Info | Homo sapiens | [3] | |||
| Fatty acid desaturase 2 (FADS2) | DME Info | Homo sapiens | [3] | |||
| Iduronate 2-sulfatase (IDS) | DME Info | Homo sapiens | [3] | |||
| Phosphorylethanolamine transferase (PCYT2) | DME Info | Homo sapiens | [2], [6] | |||
| Serum paraoxonase/arylesterase 1 (PON1) | DME Info | Homo sapiens | [3] | |||
| Thymidylate synthase (TYMS) | DME Info | Homo sapiens | [7] | |||
| UDP-glucuronosyltransferase 1A10 (UGT1A10) | DME Info | Homo sapiens | [2], [6] | |||
| UDP-glucuronosyltransferase 1A8 (UGT1A8) | DME Info | Homo sapiens | [3] | |||
| DME(s) Induced by This Xenobiotics | ||||||
| Aldo-keto reductase 1C1 (AKR1C1) | DME Info | Homo sapiens | [2] | |||
| Butyrylcholine esterase (BCHE) | DME Info | Homo sapiens | [7], [5] | |||
| Folylpolyglutamate synthase (FPGS) | DME Info | Homo sapiens | [4] | |||
| Gamma-Glu-X carboxypeptidase (GGH) | DME Info | Homo sapiens | [5] | |||
| L-glutamine amidohydrolase (GLS) | DME Info | Homo sapiens | [3] | |||
| Histamine N-methyltransferase (HNMT) | DME Info | Homo sapiens | [7] | |||
| Purine nucleoside phosphorylase (PNP) | DME Info | Homo sapiens | [8] | |||
| Prostaglandin reductase 1 (PTGR1) | DME Info | Homo sapiens | [7] | |||
| Putrescine acetyltransferase (SSAT1) | DME Info | Homo sapiens | [2], [5] | |||
| Thiopurine methyltransferase (TPMT) | DME Info | Homo sapiens | [1] | |||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

