Details of the Drug Metabolized by Drug-Metabolizing Enzyme (DME)
| General Information of Drug (ID: DR1189) | ||||||
|---|---|---|---|---|---|---|
| Drug Name |
Oleandomycin
|
|||||
| Synonyms |
Oleandomycin A; Amimycin; Landomycin; Matromycin; Antibiotic PA 105; LMPK04000007; P8ZQ646136; PA 105; Prestwick3_000152; RZPAKFUAFGMUPI-QESOVKLGSA-N; Romicil; SCHEMBL3717; ZINC85432018; oleandomycin; 3922-90-5; AB00513809; AC1L2I65; BDBM234401; BPBio1_000314; BSPBio_000284; C01946; CHEBI:16869; CHEMBL606258; CS-0063452; E704; HY-116010; NCGC00179617-01; UNII-P8ZQ646136
|
|||||
| Indication | Acute upper respiratory infection [ICD11: CA07] | Phase 4 | [1] | |||
| Structure |
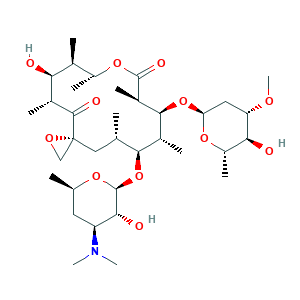 |
|||||
| 3D MOL | 2D MOL | |||||
| Pharmaceutical Properties | Molecular Weight | 687.9 | Topological Polar Surface Area | 166 | ||
| Heavy Atom Count | 48 | Rotatable Bond Count | 6 | |||
| Hydrogen Bond Donor Count | 3 | Hydrogen Bond Acceptor Count | 13 | |||
| Cross-matching ID |
|
|||||
| The Metabolic Roadmap of This Drug | |||||
|---|---|---|---|---|---|
| The Full List of Drug Metabolites (DM) of This Drug | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||
| The Full List of Metabolic Reaction (MR) of This Drug | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||
| Drug-Metabolizing Enzyme(s) (DME) Metabolizing This Drug | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

