| General Information of Drug (ID:
DR0722) |
| Drug Name |
Fluphenazine decanoate
|
| Prodrug Info |
Fluphenazine decanoate is the prodrug of Fluphenazine sulfoxide
|
| Synonyms |
Flufenazin; Flufenazina; Flufenazina [DCIT]; Fluorfenazine; Fluorophenazine; Fluorphenazine; Fluphenazine [INN:BAN]; Fluphenazinum; Fluphenazinum [INN-Latin]; Ftorphenazine; Moditen (Tabl or elixir); Pacinol; Phthorphenazine; Prolixin; SQ 4918; Sevinol; Siqualine; Siqualon; Triflumethazine; Vespazine; Yespazine; 10-(3-(2-Hydroxyethyl)piperazinopropyl)-2-(trifluoromethyl)phenothiazine; 2-(4-(3-(2-(trifluoromethyl)-10H-phenothiazin-10-yl)propyl)piperazin-1-yl)ethanol; 69-23-8; HSDB 3334; UNII-S79426A41Z; Dapotum; Elinol; FLUPHENAZINE; Dapotum D; FMU62K1L3C; Flufenazine decanoate; Fluorophenazine decanoate; Fluphenaline decanoate; Fluphenazine O-decanoate; Fluphenazine decanoate; Fluphenazine depot; Modecate; Moditen depot; Prolixin decanoate; SQ 10,733; 2-(4-(3-(2-(Trifluoromethyl)phenothiazin-10-yl)propyl)-1-piperazinyl)ethyl decanoate; 2-[4-[3-[2-(trifluoromethyl)phenothiazin-10-yl]propyl]piperazin-1-yl]ethyl decanoate; 5002-47-1; BRN 0599852; C32H44F3N3O2S; CCRIS 3954; EINECS 225-672-4; NSC 169510; UNII-FMU62K1L3C
|
| Indication |
Schizophrenia
[ICD11: 6A20]
|
Approved
|
[1]
|
| Structure |
|
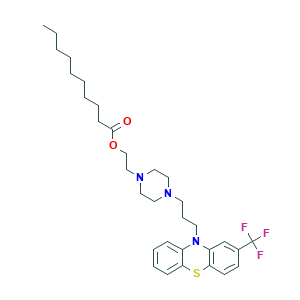 |
|
3D MOL is unavailable
|
2D MOL
|
| Pharmaceutical Properties |
Molecular Weight |
591.8 |
Topological Polar Surface Area |
61.3 |
| Heavy Atom Count |
41 |
Rotatable Bond Count |
16 |
| Hydrogen Bond Donor Count |
0 |
Hydrogen Bond Acceptor Count |
9 |
| Cross-matching ID |
- PubChem CID
- 3388
- PubChem SID
-
10157
; 442284
; 5367841
; 7847858
; 8152159
; 16865942
; 29222523
; 48416028
; 49960438
; 57321771
; 75108673
; 88771022
; 103770837
; 103821152
; 104303368
; 108136249
; 117505029
; 126674203
; 129480527
; 134223143
; 134338575
; 134338624
; 134986213
; 141522722
; 142679208
; 144227482
; 162172916
; 163134728
; 175266897
; 179225767
; 221672973
; 223671476
; 223704746
; 226426349
; 241102944
- ChEBI ID
-
- CAS Number
-
- TTD Drug ID
- D03NTJ
- Formula
- C32H44F3N3O2S
- Canonical SMILES
- CCCCCCCCCC(=O)OCCN1CCN(CC1)CCCN2C3=CC=CC=C3SC4=C2C=C(C=C4)C(F)(F)F
- InChI
- 1S/C32H44F3N3O2S/c1-2-3-4-5-6-7-8-14-31(39)40-24-23-37-21-19-36(20-22-37)17-11-18-38-27-12-9-10-13-29(27)41-30-16-15-26(25-28(30)38)32(33,34)35/h9-10,12-13,15-16,25H,2-8,11,14,17-24H2,1H3
- InChIKey
- VIQCGTZFEYDQMR-UHFFFAOYSA-N
|
|
|
|
|
|
|
|
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.


