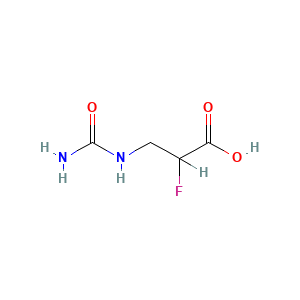
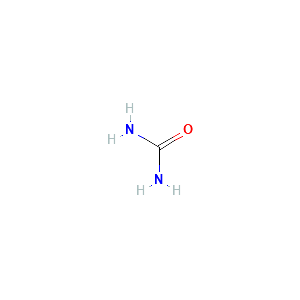
| References |
| 1 |
DrugBank(Pharmacology-Metabolism):Tegafur-uracil
|
| 2 |
A new, validated HPLC-MS/MS method for the simultaneous determination of the anti-cancer agent capecitabine and its metabolites: 5'-deoxy-5-fluorocytidine, 5'-deoxy-5-fluorouridine, 5-fluorouracil and 5-fluorodihydrouracil, in human plasma
|
| 3 |
Metabolism and function in animal tissues of agmatine, a biogenic amine formed from arginine Amino Acids. 2004 Feb;26(1):3-8. doi: 10.1007/s00726-003-0030-z.
|
| 4 |
Ammonia Transporters and Their Role in Acid-Base Balance Physiol Rev. 2017 Apr;97(2):465-494. doi: 10.1152/physrev.00011.2016.
|
| 5 |
Comparative misonidazole metabolism in anaerobic bacteria and hypoxic Chinese hamster lung fibroblast (V-79-473) cells
|
| 6 |
A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. Nat Commun. 2021 Aug 26;12(1):5150. doi: 10.1038/s41467-021-25369-x.
|
| 7 |
Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism
|