Details of the Drug Metabolized by Drug-Metabolizing Enzyme (DME)
| General Information of Drug (ID: DR1072) | ||||||
|---|---|---|---|---|---|---|
| Drug Name |
Metoprolol tartrate
|
|||||
| Synonyms |
Metoproferm; Metoprolin; Metoprolol AL; Metoprolol Atid; Metoprolol Basics; Metoprolol PB; Metoprolol Stada; Metoprolol Verla; Metoprolol acis; Metoprolol tartrate; Metoprolol-GRY; Metoprolol-Wolf; Metoprolol-rpm; Prelis; Proken M; Prolaken; Ritmolol; Selectadril; Selokeen; Selopral; Sigaprolol; Slow lopresor; Slow-Lopresor; Topromel; Vasocardin; 56392-17-7; Beloc; Lanoc; Metop; Minax; Apo-Metoprolol; Arbralene; Azumetop; Beprolo; Bloksan; Bloxan; Cardoxone; Jeprolol; Kapodine; Mepolol; Meto-Isis; Metoberag; Metoberta; Metocard; Metolol; Metomerck
|
|||||
| Indication | Angina pectoris [ICD11: BA40] | Approved | [1] | |||
| Structure |
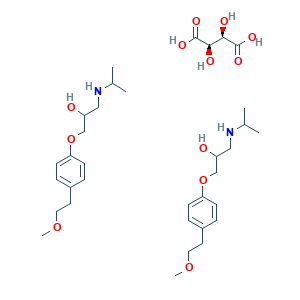 |
|||||
| 3D MOL is unavailable | 2D MOL | |||||
| Pharmaceutical Properties | Molecular Weight | 684.8 | Topological Polar Surface Area | 217 | ||
| Heavy Atom Count | 48 | Rotatable Bond Count | 21 | |||
| Hydrogen Bond Donor Count | 8 | Hydrogen Bond Acceptor Count | 14 | |||
| Cross-matching ID |
|
|||||
| The Metabolic Roadmap of This Drug | |||||
|---|---|---|---|---|---|
| The Full List of Drug Metabolites (DM) of This Drug | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| The Full List of Metabolic Reaction (MR) of This Drug | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Drug-Metabolizing Enzyme(s) (DME) Metabolizing This Drug | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

