Details of the Drug Metabolized by Drug-Metabolizing Enzyme (DME)
| General Information of Drug (ID: DR2669) | ||||||
|---|---|---|---|---|---|---|
| Drug Name |
Ginsenoside Rb1
|
|||||
| Synonyms |
Arasaponin E1; Ginsenoside rb1; Gynosaponin C; Panax saponin E; Panaxsaponin E; Pseudoginsenoside D; Sanchinoside E1; ginsenoside-Rb1; (20S)-ginsenoside Rb1; 2-O-beta-Glucopyranosyl-(3beta,12beta)-20-((6-O-beta-D-glucopyranosyl-beta-D-glucopyranosyl)oxy)-12-hydroxydammar-24-en-3-yl-beta-D-glucopyranoside; 20(S)-ginsenoside Rb1; 41753-43-9; 7413S0WMH6; BIDD:ER0108; C54H92O23; CHEBI:67989; CHEMBL501515; GRb 1; GS-Rb1; GSRb1; GZYPWOGIYAIIPV-JBDTYSNRSA-N; Gypenoside III; NSC 310103; UNII-7413S0WMH6
|
|||||
| Indication | Obesity [ICD11: 5B81] | Investigative | [1] | |||
| Structure |
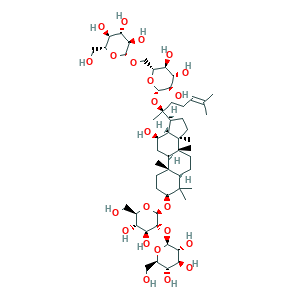 |
|||||
| 3D MOL is unavailable | 2D MOL | |||||
| Pharmaceutical Properties | Molecular Weight | 1109.3 | Topological Polar Surface Area | 377 | ||
| Heavy Atom Count | 77 | Rotatable Bond Count | 16 | |||
| Hydrogen Bond Donor Count | 15 | Hydrogen Bond Acceptor Count | 23 | |||
| Cross-matching ID |
|
|||||
| The Metabolic Roadmap of This Drug | |||||
|---|---|---|---|---|---|
| The Full List of Drug Metabolites (DM) of This Drug | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||
| The Full List of Metabolic Reaction (MR) of This Drug | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||
| Drug-Metabolizing Enzyme(s) (DME) Metabolizing This Drug | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Enzyme Kinetic Data of This Drug | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

