Detail Information of Xenobiotics
| General Information of Xenobiotics (ID: XEO00349) | ||||||
|---|---|---|---|---|---|---|
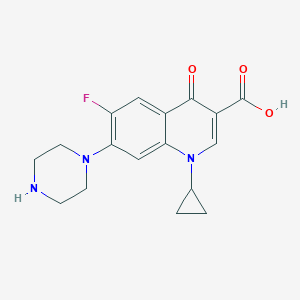
| Xenobiotics Name |
Ciprofloxacin
|
|||||
| Xenobiotics Type |
Pharmaceutical Agent(s)
|
|||||
| Classification |
Approved/Marketed Drug
|
|||||
| Structure |
<iframe style="width: 300px; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=2764"></iframe>
|
 |
||||
| 3D MOL | 2D MOL | |||||
| PubChem CID | ||||||
| DME(s) Modulated by This Xenobiotics | ||||||
| DME(s) Inhibited by This Xenobiotics | ||||||
| Aminoglycoside acetyltransferase (aac) | DME Info | Providencia stuartii | [1] | |||
| Beta-lactamase (blaB) | DME Info | Morganella morganii | [1] | |||
| Aminoglycoside O-phosphotransferase (aphA6) | DME Info | Providencia stuartii | [1] | |||
| Azoreductase (azoR) | DME Info | Escherichia coli | [1] | |||
| L,D-carboxypeptidase A (ldcA) | DME Info | Escherichia coli | [1] | |||
| Beta-lactamase (blaB) | DME Info | Escherichia coli | [1] | |||
| Cytidine deaminase (cdd) | DME Info | Escherichia coli | [2] | |||
| Cytidine deaminase (cdd) | DME Info | Klebsiella pneumoniae | [2] | |||
| Chloramphenicolase (chlR) | DME Info | Escherichia coli | [1] | |||
| Glycoside hydrolase (cscA) | DME Info | Escherichia coli | [1] | |||
| NADPH-dependent curcumin reductase (curA) | DME Info | Escherichia coli | [1] | |||
| Cytochrome P450 1A2 (CYP1A2) | DME Info | Homo sapiens | [3] | |||
| Cytochrome P450 3A4 (CYP3A4) | DME Info | Homo sapiens | [4] | |||
| Unclear metabolic mechanism (DME-unclear) | DME Info | Escherichia coli | [1] | |||
| Unclear metabolic mechanism (DME-unclear) | DME Info | Providencia stuartii | [1] | |||
| Glutamate decarboxylase (gadB) | DME Info | Escherichia coli | [1] | |||
| D-Lactate dehydrogenase (ldhA) | DME Info | Escherichia coli | [1] | |||
| D-Lactate dehydrogenase (ldhA) | DME Info | Pseudomonas aeruginosa | [5] | |||
| Molybdopterin-dependent enzyme (molD) | DME Info | Escherichia coli | [1] | |||
| Glutamate racemase (MurI) | DME Info | Escherichia coli | [1] | |||
| Arylamine N-acetyltransferase (NAT) | DME Info | Morganella morganii | [1] | |||
| Arylamine N-acetyltransferase (NAT) | DME Info | Pseudomonas aeruginosa | [5] | |||
| N-ethylmaleimide reductase (nemA) | DME Info | Escherichia coli | [1] | |||
| NADPH-dependent oxidoreductase (nfrA) | DME Info | Staphylococcus aureus | [6] | |||
| Oxygen-insensitive NADPH nitroreductase A (nfsA) | DME Info | Escherichia coli | [1] | |||
| Oxygen-insensitive NADPH nitroreductase B (nfsB) | DME Info | Escherichia coli | [1] | |||
| NADH dehydrogenase (nuoE) | DME Info | Streptomyces griseus | [1] | |||
| Hydroxybenzoate 3-monooxygenase (pobA) | DME Info | Pseudomonas aeruginosa | [5] | |||
| Serum paraoxonase/arylesterase 1 (PON1) | DME Info | Homo sapiens | [7] | |||
| Tyramine oxidase (tynA) | DME Info | Escherichia coli | [1] | |||
| Beta-glucuronidase (uidA) | DME Info | Escherichia coli | [2] | |||
| DME(s) Induced by This Xenobiotics | ||||||
| Aldehyde dehydrogenase 1 (ALDH1) | DME Info | Homo sapiens | [8] | |||
| Xenobiotics-DME Activity Data | ||||||
| Xenobiotics-DME Activity Data | Cytochrome P450 3A4 (CYP3A4) | DME Info | IC50 > 10 microM | [4] | ||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

