Details of Drug-Metabolizing Enzyme (DME)
| Full List of Drug(s) Metabolized by This DME | |||||
|---|---|---|---|---|---|
| Drugs Approved by FDA | Click to Show/Hide the Full List of Drugs: 287 Drugs | ||||
Bambuterol |
Drug Info | Approved | Asthma | ICD11: CA23 | [1] |
Cobicistat |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [2] |
Bupropion |
Drug Info | Approved | Nicotine dependence | ICD11: 6C4A | [3] |
Tamoxifen citrate |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [4] |
Yn-968D1 |
Drug Info | Approved | Breast cancer | ICD11: 2C60-2C6Y | [5] |
Erlotinib hydrochloride |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [6] |
Idelalisib |
Drug Info | Approved | Chronic lymphocytic leukaemia | ICD11: 2A82 | [7] |
Estrone |
Drug Info | Approved | Menopausal disorder | ICD11: GA30 | [8] |
Methylphenidate |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [9] |
Nicotine |
Drug Info | Approved | Nicotine dependence | ICD11: 6C4A | [10] |
Cocaine |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [11] |
Pimavanserin |
Drug Info | Approved | Parkinsonism | ICD11: 8A00 | [12] |
Bortezomib |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [13] |
Delavirdine mesylate |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [14] |
Hydrocodone |
Drug Info | Approved | Pain | ICD11: MG30-MG3Z | [15] |
Pindolol |
Drug Info | Approved | Hypertension | ICD11: BA00-BA04 | [16] |
Antazoline |
Drug Info | Approved | Nasal congestion | ICD11: MD11 | [17] |
Galantamine hydrobromide |
Drug Info | Approved | Alzheimer disease | ICD11: 8A20 | [18] |
Ibrutinib |
Drug Info | Approved | Mantle cell lymphoma | ICD11: 2A85 | [19] |
Ketoconazole |
Drug Info | Approved | Dermatophytosis | ICD11: 1F28 | [20] |
Metoclopramide hydrochloride |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [21] |
Primaquine |
Drug Info | Approved | Malaria | ICD11: 1F40 | [22] |
Progesterone |
Drug Info | Approved | Premature labour | ICD11: JB00 | [23] |
Almogran |
Drug Info | Approved | Migraine | ICD11: 8A80 | [24] |
Almogran |
Drug Info | Approved | Migraine | ICD11: 8A80 | [25], [26] |
Imatinib mesylate |
Drug Info | Approved | Chronic myelogenous leukaemia | ICD11: 2A20 | [13] |
Levobunolol |
Drug Info | Approved | Glaucoma | ICD11: 9C61 | [27] |
Levodopa |
Drug Info | Approved | Parkinsonism | ICD11: 8A00 | [28] |
Lidocaine |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [29] |
Cannabidiol |
Drug Info | Approved | Lennox-Gastaut syndrome | ICD11: 8A62 | [30] |
Capsaicin |
Drug Info | Approved | Herpes zoster | ICD11: 1E91 | [31] |
Clomipramine hydrochloride |
Drug Info | Approved | Obsessive-compulsive disorder | ICD11: 6B20 | [32] |
Doxazosin |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [33] |
Lapatinib ditosylate |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [6] |
Pitavastatin |
Drug Info | Approved | Hypertriglyceridaemia | ICD11: 5C80 | [34] |
Ticlopidine |
Drug Info | Approved | Cerebral stroke | ICD11: 8B11 | [35] |
Tipranavir |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [36] |
Almotriptan malate |
Drug Info | Approved | Migraine | ICD11: 8A80 | [25] |
Asenapine maleate |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [37] |
Betaxolol |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [38] |
Bupivacaine hydrochloride |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [39] |
Clomipramine |
Drug Info | Approved | Depression | ICD11: 6A70-6A7Z | [40] |
Clomipramine |
Drug Info | Approved | Depression | ICD11: 6A70-6A7Z | [32], [41], [42] |
Cyclobenzaprine |
Drug Info | Approved | Depression | ICD11: 6A71 | [43] |
Duloxetine hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [44] |
Labetalol |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [45] |
Mirabegron |
Drug Info | Approved | Overactive bladder | ICD11: GC50 | [46] |
Oxycodone hydrochloride |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [47] |
Oxymorphone |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [48] |
Ranolazine |
Drug Info | Approved | Angina pectoris | ICD11: BA40 | [49] |
Tacrine hydrochloride |
Drug Info | Approved | Alzheimer disease | ICD11: 8A20 | [24] |
Trabectedin |
Drug Info | Approved | Leiomyosarcoma | ICD11: 2B58 | [50] |
Clomiphene citrate |
Drug Info | Approved | Female infertility | ICD11: GA31 | [51] |
Clomiphene citrate |
Drug Info | Approved | Female infertility | ICD11: GA31 | [52] |
Dopamine hydrochloride |
Drug Info | Approved | Parkinsonism | ICD11: 8A00 | [53] |
Fluvoxamine maleate |
Drug Info | Approved | Obsessive-compulsive disorder | ICD11: 6B20 | [54] |
Halothane |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [55] |
Hydrocodone bitartrate |
Drug Info | Approved | Neuropathic pain | ICD11: 8E43 | [56] |
Indinavir sulfate |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [57] |
Panobinostat lactate |
Drug Info | Approved | Multiple myeloma | ICD11: 2A83 | [58] |
Quinine sulfate |
Drug Info | Approved | Malaria | ICD11: 1F40 | [59] |
Remdesivir |
Drug Info | Approved | Coronavirus Disease 2019 (COVID-19) | ICD11: 1D6Y | [60] |
Tamsulosin |
Drug Info | Approved | Prostatic hyperplasia | ICD11: GA90 | [61] |
Tamsulosin |
Drug Info | Approved | Prostatic hyperplasia | ICD11: GA90 | [62] |
Brexpiprazole |
Drug Info | Approved | Bipolar disorder | ICD11: 6A60 | [63] |
Caffeine |
Drug Info | Approved | Orthostatic hypotension | ICD11: BA21 | [64] |
Carvedilol |
Drug Info | Approved | Congestive heart failure | ICD11: BD10 | [13] |
Deutetrabenazine |
Drug Info | Approved | Huntington disease | ICD11: 8A01 | [65] |
Dextromethorphan hydrobromide |
Drug Info | Approved | Atherosclerosis | ICD11: BA80 | [66] |
Doxepin hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [67] |
Eliglustat tartrate |
Drug Info | Approved | Gaucher disease | ICD11: 5C56 | [68] |
Formoterol |
Drug Info | Approved | Asthma | ICD11: CA23 | [69] |
Haloperidol decanoate |
Drug Info | Approved | Agitation/aggression | ICD11: 6D86 | [70] |
Lasofoxifene |
Drug Info | Approved | Osteoporosis | ICD11: FB83 | [71] |
Lasofoxifene |
Drug Info | Approved | Osteoporosis | ICD11: FB83 | [72] |
Lorcaserin |
Drug Info | Approved | Obesity | ICD11: 5B81 | [73] |
Nateglinide |
Drug Info | Approved | Diabetes mellitus | ICD11: 5A10 | [74] |
Nevirapine |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [75] |
Ondansetron |
Drug Info | Approved | Gastritis | ICD11: DA42 | [76] |
Pomalidomide |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [77] |
Propafenone hydrochloride |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [78] |
Propranolol hydrochloride |
Drug Info | Approved | Migraine | ICD11: 8A80 | [79] |
Trazodone hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [80] |
Umeclidinium bromide |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [81] |
Amitriptyline hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [82] |
Amprenavir |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [83] |
Artemether |
Drug Info | Approved | Malaria | ICD11: 1F40 | [84] |
Atomoxetine hydrochloride |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [85] |
Benzatropine |
Drug Info | Approved | Agitation/aggression | ICD11: 6D86 | [86] |
Betrixaban |
Drug Info | Approved | Venous thromboembolism | ICD11: BD72 | [87] |
Bromfenac |
Drug Info | Approved | Cataract | ICD11: 9B10 | [88] |
Buspirone |
Drug Info | Approved | Anxiety disorder | ICD11: 6B00 | [89] |
Captopril |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [55] |
Cevimeline hydrochloride |
Drug Info | Approved | Sjogren syndrome | ICD11: 4A43 | [90] |
Cinacalcet hydrochloride |
Drug Info | Approved | Hyperparathyroidism | ICD11: 5A51 | [91] |
Codeine |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [66] |
Darifenacin hydrobromide |
Drug Info | Approved | Overactive bladder | ICD11: GC50 | [92] |
Desvenlafaxine succinate |
Drug Info | Approved | Depression | ICD11: 6A71 | [93] |
Diltiazem hydrochloride |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [94] |
Donepezil hydrochloride |
Drug Info | Approved | Alzheimer disease | ICD11: 8A20 | [95] |
Doxorubicin |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [96] |
Dronedarone hydrochloride |
Drug Info | Approved | Atrial fibrillation | ICD11: BC81 | [97] |
Exjade |
Drug Info | Approved | Hyperphosphatemia | ICD11: 5C64 | [98] |
Fluoxetine hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [99] |
Fluvastatin sodium |
Drug Info | Approved | Hypertriglyceridaemia | ICD11: 5C80 | [100] |
Imipramine hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [101] |
Indomethacin |
Drug Info | Approved | Rheumatoid arthritis | ICD11: FA20 | [102] |
Istradefylline |
Drug Info | Approved | Parkinsonism | ICD11: 8A00 | [103] |
Lesinurad |
Drug Info | Approved | Uncontrolled gout | ICD11: FA25 | [104] |
Letermovir |
Drug Info | Approved | Cytomegalovirus infection | ICD11: 1D82 | [105] |
Levomepromazine |
Drug Info | Approved | Insomnia | ICD11: 7A00 | [106] |
Lisdexamfetamine |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [107] |
Metoprolol succinate |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [108] |
Metoprolol tartrate |
Drug Info | Approved | Angina pectoris | ICD11: BA40 | [108] |
Morphine |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [109] |
Nefazodone |
Drug Info | Approved | Depression | ICD11: 6A71 | [110] |
Olanzapine |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [111] |
Phenytoin |
Drug Info | Approved | Epilepsy | ICD11: 8A60 | [40] |
Quinidine |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [112] |
Revefenacin |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [113] |
Ropivacaine hydrochloride |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [55] |
Rucaparib |
Drug Info | Approved | Ovarian cancer | ICD11: 2C73 | [114] |
Simvastatin |
Drug Info | Approved | Dyslipidaemia | ICD11: 5C81 | [115] |
Tapentadol |
Drug Info | Approved | Neuropathic pain | ICD11: 8E43 | [116] |
Tegaserod |
Drug Info | Approved | Irritable bowel syndrome | ICD11: DD91 | [117] |
Zolpidem tartrate |
Drug Info | Approved | Insomnia | ICD11: 7A00 | [118] |
Amiodarone hydrochloride |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [119] |
Amoxapine |
Drug Info | Approved | Depression | ICD11: 6A71 | [120] |
Azelastine |
Drug Info | Approved | Allergic rhinitis | ICD11: CA08 | [121] |
Buprenorphine hydrochloride |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [122] |
Chlorpromazine hydrochloride |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [70] |
Citalopram hydrobromide |
Drug Info | Approved | Depression | ICD11: 6A71 | [123] |
Clonidine |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [124] |
Clozapine |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [125] |
Diphenhydramine |
Drug Info | Approved | Meniere disease | ICD11: AB31 | [126] |
Dolasetron mesylate |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [127] |
Ethosuximide |
Drug Info | Approved | Epilepsy | ICD11: 8A60 | [128] |
Fingolimod |
Drug Info | Approved | Multiple sclerosis | ICD11: 8A40 | [129] |
Fingolimod hydrochloride |
Drug Info | Approved | Multiple sclerosis | ICD11: 8A40 | [129] |
Loxapine succinate |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [130] |
Metamfetamine |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [131] |
Methamphetamine hydrochloride |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [131] |
Mirtazapine |
Drug Info | Approved | Depression | ICD11: 6A71 | [132] |
Nalbuphine |
Drug Info | Approved | Pain | ICD11: MG30-MG3Z | [133] |
Oxycodone |
Drug Info | Approved | Pain | ICD11: MG30-MG3Z | [134] |
Paroxetine hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [135] |
Pazopanib |
Drug Info | Approved | Renal cell carcinoma | ICD11: 2C90 | [136] |
Perphenazine |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [70] |
Pimozide |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [137] |
Ponatinib hydrochloride |
Drug Info | Approved | Acute lymphoblastic leukemia | ICD11: 2B33 | [138] |
Pralsetinib |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [139] |
Pralsetinib |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [140] |
Prochlorperazine |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [141] |
Propofol |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [142] |
Quetiapine fumarate |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [143] |
Ritonavir |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [144] |
Solifenacin succinate |
Drug Info | Approved | Overactive bladder | ICD11: GC50 | [145] |
Sumatriptan |
Drug Info | Approved | Migraine | ICD11: 8A80 | [146] |
Tandospirone |
Drug Info | Approved | Anxiety disorder | ICD11: 6B00-6B0Z | [147] |
Tasimelteon |
Drug Info | Approved | Insomnia | ICD11: 7A00 | [148] |
Theophylline |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [149] |
Thioridazine |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [150] |
Tramadol hydrochloride |
Drug Info | Approved | Neuropathic pain | ICD11: 8E43 | [151] |
Triclosan |
Drug Info | Approved | Malaria | ICD11: 1F40-1F45 | [152] |
Venlafaxine hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [153] |
Vilazodone hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [154] |
Vortioxetine hydrobromide |
Drug Info | Approved | Depression | ICD11: 6A71 | [155] |
Acetaminophen |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [156] |
Amphetamine |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [157] |
Aripiprazole lauroxil |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [158] |
Benzocaine |
Drug Info | Approved | Anaesthesia | ICD11: 9A76-9A78 | [159] |
Binimetinib |
Drug Info | Approved | Melanoma | ICD11: 2C30 | [160] |
Chlorpheniramine |
Drug Info | Approved | Allergic rhinitis | ICD11: CA08 | [161] |
Ciclesonide |
Drug Info | Approved | Asthma | ICD11: CA23 | [162] |
Dapagliflozin |
Drug Info | Approved | Diabetes mellitus | ICD11: 5A10 | [163] |
Desloratadine |
Drug Info | Approved | Allergic rhinitis | ICD11: CA08 | [164] |
Dutasteride |
Drug Info | Approved | Prostatic hyperplasia | ICD11: GA90 | [44] |
Fesoterodine |
Drug Info | Approved | Overactive bladder | ICD11: GC50 | [165] |
Gefitinib |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [166] |
Iloperidone |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [167] |
Levallorphan |
Drug Info | Approved | Narcotic depression | ICD11: 6A70-6A7Z | [168] |
Levomethadyl acetate hydrochloride |
Drug Info | Approved | Opiate dependence | ICD11: 6C43 | [169] |
Levomilnacipran |
Drug Info | Approved | Depression | ICD11: 6A71 | [170] |
Loperamide hydrochloride |
Drug Info | Approved | Irritable bowel syndrome | ICD11: DD91 | [171] |
Lovastatin |
Drug Info | Approved | Hypertriglyceridaemia | ICD11: 5C80 | [172] |
Maprotiline hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [173] |
Meclizine |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [174] |
Methadone |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [175] |
Methyldopa |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [176] |
Mexiletine hydrochloride |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [177] |
Midodrine |
Drug Info | Approved | Orthostatic hypotension | ICD11: BA21 | [178] |
Palonosetron hydrochloride |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [76] |
Pitolisant |
Drug Info | Approved | Excessive daytime sleepiness | ICD11: MG42 | [179] |
Procainamide hydrochloride |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [180] |
Promethazine |
Drug Info | Approved | Functional nausea/vomiting | ICD11: DD90 | [178] |
Ranitidine |
Drug Info | Approved | Peptic ulcer | ICD11: DA61 | [181] |
Tapinarof |
Drug Info | Approved | Discovery agent | ICD: N.A. | [182] |
Timolol maleate |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [183] |
Tolterodine tartrate |
Drug Info | Approved | Overactive bladder | ICD11: GC50 | [184] |
Vinorelbine tartrate |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [96] |
Zidovudine |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [55], [185] |
Acebutolol |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [27] |
Alogliptin |
Drug Info | Approved | Diabetes mellitus | ICD11: 5A10 | [186] |
Arformoterol |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [187] |
Azilsartan |
Drug Info | Approved | Hypertension | ICD11: BA00-BA04 | [188], [189] |
Cariprazine hydrochloride |
Drug Info | Approved | Bipolar disorder | ICD11: 6A60 | [190] |
Carteolol hydrochloride |
Drug Info | Approved | Glaucoma | ICD11: 9C61 | [191] |
Celecoxib |
Drug Info | Approved | Rheumatoid arthritis | ICD11: FA20 | [192] |
Cilostazol |
Drug Info | Approved | Arteriosclerosis obliterans | ICD11: BD40 | [193] |
Clofibrate |
Drug Info | Approved | Hypertriglyceridaemia | ICD11: 5C80 | [194] |
Dexchlorpheniramine maleate |
Drug Info | Approved | Rheumatoid arthritis | ICD11: FA20 | [161] |
Dextroamphetamine |
Drug Info | Approved | Narcolepsy | ICD11: 7A20 | [157] |
Eletriptan hydrobromide |
Drug Info | Approved | Migraine | ICD11: 8A80 | [195] |
Escitalopram |
Drug Info | Approved | Depression | ICD11: 6A71 | [196] |
Esmolol |
Drug Info | Approved | Atrial fibrillation | ICD11: BC81 | [27] |
Flecainide acetate |
Drug Info | Approved | Ventricular tachyarrhythmia | ICD11: BC71 | [197] |
Flibanserin |
Drug Info | Approved | Inhibited sexual desire | ICD11: HA00 | [198] |
Fluvoxamine |
Drug Info | Approved | Depression | ICD11: 6A70-6A7Z | [199] |
Hydromorphone |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [200] |
Iclaprim |
Drug Info | Approved | Methicillin-resistant staphylococcus infection | ICD11: 1D01 | [201] |
Loratadine |
Drug Info | Approved | Allergic rhinitis | ICD11: CA08 | [202] |
Mephenytoin |
Drug Info | Approved | Epilepsy | ICD11: 8A60 | [203] |
Nicardipine hydrochloride |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [204] |
Nifedipine |
Drug Info | Approved | Angina pectoris | ICD11: BA40 | [205] |
Oxamniquine |
Drug Info | Approved | Schistosomiasis | ICD11: 1F86 | [206] |
Paliperidone |
Drug Info | Approved | Bipolar disorder | ICD11: 6A60 | [207] |
Penbutolol |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [27] |
Pentamidine isethionate |
Drug Info | Approved | Pneumocystis pneumonia | ICD11: CA40 | [22] |
Phenmetrazine |
Drug Info | Approved | Obesity | ICD11: 5B80-5B81 | [208] |
Selegiline |
Drug Info | Approved | Major depressive disorder | ICD11: 6A70-6A7Z | [209] |
Selegiline hydrochloride |
Drug Info | Approved | Parkinsonism | ICD11: 8A00 | [210] |
Sertraline hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [211] |
Sildenafil citrate |
Drug Info | Approved | Erectile dysfunction | ICD11: HA01 | [13] |
Tetrabenazine |
Drug Info | Approved | Huntington disease | ICD11: 8A01 | [212] |
Tiotropium |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [213] |
Tiotropium Bromide |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [214] |
Triclabendazole |
Drug Info | Approved | Fascioliasis | ICD11: 1F82 | [215] |
Trimipramine |
Drug Info | Approved | Depression | ICD11: 6A71 | [216] |
Ulipristal |
Drug Info | Approved | Contraception | ICD11: JA65 | [217] |
Umeclidinium |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [218] |
Umeclidinium |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [219] |
Valbenazine |
Drug Info | Approved | Dyskinesia | ICD11: 8A02 | [220] |
Benzyl alcohol |
Drug Info | Approved | Pediculosis | ICD11: 1G00 | [221] |
Bisoprolol |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [222] |
Chloroquine |
Drug Info | Approved | Malaria | ICD11: 1F40 | [223] |
Chloroquine |
Drug Info | Approved | Malaria | ICD11: 1F40 | [224] |
Chlorzoxazone |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [225] |
Clevidipine |
Drug Info | Approved | Essential hypertension | ICD11: BA00 | [226] |
Dacomitinib |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [227] |
Desipramine |
Drug Info | Approved | Attention deficit hyperactivity disorder | ICD11: 6A05 | [228] |
Encorafenib |
Drug Info | Approved | Melanoma | ICD11: 2C30 | [229] |
Epinastine |
Drug Info | Approved | Conjunctivitis | ICD11: 9A60 | [230] |
Fluphenazine decanoate |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [125] |
Fluphenazine enanthate |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [125] |
Glycopyrrolate |
Drug Info | Approved | Chronic obstructive pulmonary disease | ICD11: CA22 | [231] |
Halofantrine |
Drug Info | Approved | Malaria | ICD11: 1F40 | [223] |
Hydrogen peroxide |
Drug Info | Approved | Emphysema | ICD11: CA21 | [232] |
Ibudilast |
Drug Info | Approved | Castleman's disease | ICD11: 4B2Y | [233] |
Idarubicin |
Drug Info | Approved | Acute myeloid leukaemia | ICD11: 2A60 | [234] |
Methyprylon |
Drug Info | Approved | Insomnia | ICD11: 7A00 | [235] |
Metipranolol |
Drug Info | Approved | Glaucoma | ICD11: 9C61 | [27] |
Nebivolol hydrochloride |
Drug Info | Approved | Pulmonary hypertension | ICD11: BB01 | [236] |
Nortriptyline hydrochloride |
Drug Info | Approved | Depression | ICD11: 6A71 | [237] |
Pachycarpine |
Drug Info | Approved | Postpartum haemorrhage | ICD11: JA43 | [238] |
Piperazine |
Drug Info | Approved | Ascariasis | ICD11: 1F62 | [239] |
Prinomastat |
Drug Info | Approved | Lung cancer | ICD11: 2C25 | [240] |
Rifampicin |
Drug Info | Approved | Osteoporosis | ICD11: FB83 | [241] |
Ripretinib |
Drug Info | Approved | Gastrointestinal stromal tumour | ICD11: 2B5B | [242] |
Risperidone |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [243] |
Sevoflurane |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [244] |
Tafenoquine |
Drug Info | Approved | Malaria | ICD11: 1F40 | [245] |
Tilidine |
Drug Info | Approved | Pain | ICD11: MG30-MG3Z | [246] |
Vinblastine sulfate |
Drug Info | Approved | Breast cancer | ICD11: 2C60 | [96] |
Zalcitabine |
Drug Info | Approved | Human immunodeficiency virus infection | ICD11: 1C60 | [247] |
Bepridil hydrochloride |
Drug Info | Approved | Angina pectoris | ICD11: BA40 | [248] |
Cefalexin |
Drug Info | Approved | Acute otitis media | ICD11: AB00 | [249] |
Elagolix sodium |
Drug Info | Approved | Endometriosis | ICD11: GA10 | [250] |
Levobetaxolol |
Drug Info | Approved | Glaucoma | ICD11: 9C61 | [27] |
Mesoridazine |
Drug Info | Approved | Schizophrenia | ICD11: 6A20 | [150] |
Methoxyflurane |
Drug Info | Approved | Anaesthesia | ICD11: 8E22 | [251] |
Rivastigmine |
Drug Info | Approved | Alzheimer disease | ICD11: 8A20 | [252] |
Sotalol |
Drug Info | Approved | Atrial fibrillation | ICD11: BC81 | [27] |
Upadacitinib |
Drug Info | Approved | Rheumatoid arthritis | ICD11: FA20 | [253] |
Upadacitinib |
Drug Info | Approved | Rheumatoid arthritis | ICD11: FA20 | [254] |
Berberine |
Drug Info | Phase 4 | Discovery agent | ICD: N.A. | [255] |
Neupro |
Drug Info | Phase 4 | Restless legs syndrome | ICD11: 7A80 | [259] |
Ingrezza |
Drug Info | Phase 4 | Dyskinesia | ICD11: 8A02 | [279] |
| Drugs in Phase 4 Clinical Trial | Click to Show/Hide the Full List of Drugs: 52 Drugs | ||||
Ajmaline |
Drug Info | Phase 4 | Atrial fibrillation | ICD11: BC81 | [55] |
Sertindole |
Drug Info | Phase 4 | Schizophrenia | ICD11: 6A20 | [256] |
Minaprine |
Drug Info | Phase 4 | Depression | ICD11: 6A71 | [257] |
Dapoxetine |
Drug Info | Phase 4 | Ejaculatory dysfunction | ICD11: HA03 | [258] |
Cinnarizine |
Drug Info | Phase 4 | Haemorrhagic stroke | ICD11: 8B20 | [13] |
Alprenolol |
Drug Info | Phase 4 | Essential hypertension | ICD11: BA00 | [106] |
Bevantolol |
Drug Info | Phase 4 | Angina pectoris | ICD11: BA40 | [27] |
Cibenzoline |
Drug Info | Phase 4 | Atrial fibrillation | ICD11: BC81 | [260] |
Mianserin |
Drug Info | Phase 4 | Depression | ICD11: 6A71 | [261] |
Remoxipride |
Drug Info | Phase 4 | Schizophrenia | ICD11: 6A20 | [55] |
Tropisetron |
Drug Info | Phase 4 | Anaesthesia | ICD11: 8E22 | [76] |
Vonoprazan |
Drug Info | Phase 4 | Gastro-oesophageal reflux disease | ICD11: DA22 | [262] |
Vonoprazan |
Drug Info | Phase 4 | Gastro-oesophageal reflux disease | ICD11: DA22 | [263] |
Zotepine |
Drug Info | Phase 4 | Schizophrenia | ICD11: 6A20 | [264] |
Astemizole |
Drug Info | Phase 4 | Allergic rhinitis | ICD11: CA08 | [265] |
Dexfenfluramine |
Drug Info | Phase 4 | Obesity | ICD11: 5B81 | [266] |
Dihydrocodeine |
Drug Info | Phase 4 | Anaesthesia | ICD11: 8E22 | [267] |
Etoricoxib |
Drug Info | Phase 4 | Anaesthesia | ICD11: 8E22 | [13] |
Idebenone |
Drug Info | Phase 4 | Mitochondrial myopathy | ICD11: 8C73 | [268] |
Imrecoxib |
Drug Info | Phase 4 | Osteoarthritis | ICD11: FA00 | [269] |
Ipecac |
Drug Info | Phase 4 | Pulmonary tuberculosis | ICD11: 1B10 | [270] |
Phenacetin |
Drug Info | Phase 4 | Anaesthesia | ICD11: 8E22 | [271] |
Terfenadine |
Drug Info | Phase 4 | Allergy | ICD11: 4A80 | [230] |
Tertatolol |
Drug Info | Phase 4 | Essential hypertension | ICD11: BA00 | [27] |
Zuclopenthixol |
Drug Info | Phase 4 | Schizophrenia | ICD11: 6A20 | [272] |
Aminophenazone |
Drug Info | Phase 4 | Anaesthesia | ICD11: 8E22 | [273] |
Chlorphenamine |
Drug Info | Phase 4 | Allergy | ICD11: 4A80 | [161] |
Dextropropoxyphene |
Drug Info | Phase 4 | Lung cancer | ICD11: 2C25 | [274] |
Encainide |
Drug Info | Phase 4 | Cardiac arrhythmia | ICD11: BC65 | [275] |
Flunarizine |
Drug Info | Phase 4 | Schizophrenia | ICD11: 6A20 | [13] |
Perazine |
Drug Info | Phase 4 | Cerebrovascular dementia | ICD11: 6D81 | [276] |
Buflomedil |
Drug Info | Phase 4 | Arterial occlusive disease | ICD11: BD4Z | [277] |
Enclomiphene |
Drug Info | Phase 4 | Female infertility | ICD11: GA31 | [278] |
Indoramin |
Drug Info | Phase 4 | Essential hypertension | ICD11: BA00 | [55] |
Lofexidine |
Drug Info | Phase 4 | Opiate dependence | ICD11: 6C43 | [280] |
Milnacipran |
Drug Info | Phase 4 | Asperger syndrome | ICD11: 6A02 | [281] |
Milnacipran |
Drug Info | Phase 4 | Asperger syndrome | ICD11: 6A02 | [282] |
Milnacipran |
Drug Info | Phase 4 | Asperger syndrome | ICD11: 6A02 | [283] |
Ramosetron |
Drug Info | Phase 4 | Irritable bowel syndrome | ICD11: DD91 | [284] |
Amsacrine |
Drug Info | Phase 4 | Acute lymphoblastic leukemia | ICD11: 2B33 | [285] |
Arotinolol |
Drug Info | Phase 4 | Essential tremor | ICD11: 8A04 | [27] |
Esmirtazapine |
Drug Info | Phase 4 | Depression | ICD11: 6A71 | [286] |
Guanoxan |
Drug Info | Phase 4 | Essential hypertension | ICD11: BA00 | [55] |
Nicergoline |
Drug Info | Phase 4 | Acne vulgaris | ICD11: ED80 | [287] |
Phenformin |
Drug Info | Phase 4 | Diabetes mellitus | ICD11: 5A10 | [11] |
Practolol |
Drug Info | Phase 4 | Cardiac arrhythmia | ICD11: BC65 | [27] |
Pyrantel |
Drug Info | Phase 4 | Hookworm disease | ICD11: 1F68 | [22] |
Aminobutyric acid |
Drug Info | Phase 4 | Epilepsy | ICD11: 8A60 | [288] |
Benzethonium |
Drug Info | Phase 4 | Methicillin-resistant staphylococcus infection | ICD11: 1D01 | [86] |
Celiprolol |
Drug Info | Phase 4 | Essential hypertension | ICD11: BA00 | [27] |
Indenolol |
Drug Info | Phase 4 | Cardiac arrhythmia | ICD11: BC65 | [27] |
Yohimbine |
Drug Info | Phase 4 | Erectile dysfunction | ICD11: HA01 | [289] |
| Drugs in Phase 3 Clinical Trial | Click to Show/Hide the Full List of Drugs: 41 Drugs | ||||
Fidarestat |
Drug Info | Phase 3 | Diabetic complication | ICD11: 5A2Y | [290] |
Fruquintinib |
Drug Info | Phase 3 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [291] |
Lisuride |
Drug Info | Phase 3 | Parkinsonism | ICD11: 8A00 | [292] |
BMS-298585 |
Drug Info | Phase 3 | Diabetes mellitus | ICD11: 5A10 | [293] |
BMS-650032 |
Drug Info | Phase 3 | Viral hepatitis | ICD11: 1E51 | [294] |
Q-100648 |
Drug Info | Phase 3 | Eclampsia | ICD11: JA25 | [27] |
Omecamtiv mecarbil |
Drug Info | Phase 3 | Heart failure | ICD11: BD10-BD1Z | [295] |
ABT-333 |
Drug Info | Phase 3 | Viral hepatitis | ICD11: 1E51 | [296] |
Chlorpromazine |
Drug Info | Phase 3 | Coronavirus Disease 2019 (COVID-19) | ICD11: 1D6Y | [297] |
DOV-220075 |
Drug Info | Phase 3 | Depression | ICD11: 6A71 | [298] |
KW-5338 |
Drug Info | Phase 3 | Gastroparesis | ICD11: DA41 | [299] |
NE-10064 |
Drug Info | Phase 3 | Cardiac arrhythmia | ICD11: BC65 | [300] |
RO-111163 |
Drug Info | Phase 3 | Hypersalivation | ICD11: DA04 | [301] |
TRYPTAMINE |
Drug Info | Phase 3 | Discovery agent | ICD: N.A. | [302] |
Bexagliflozin |
Drug Info | Phase 3 | Type-2 diabetes | ICD11: 5A11 | [303] |
CEM-102 |
Drug Info | Phase 3 | Endocarditis | ICD11: 1B12 | [304] |
GW-1000 |
Drug Info | Phase 3 | Cerebral vasospasm | ICD11: BA85 | [30] |
BMS-986165 |
Drug Info | Phase 3 | Psoriasis vulgaris | ICD11: EA90 | [305] |
LU-AE58054 |
Drug Info | Phase 3 | Alzheimer disease | ICD11: 8A20 | [306] |
Hydroxychloroquine |
Drug Info | Phase 3 | Malaria | ICD11: 1F40-1F45 | [224] |
LY-450139 |
Drug Info | Phase 3 | Alzheimer disease | ICD11: 8A20 | [307] |
R-1124 |
Drug Info | Phase 3 | Functional nausea/vomiting | ICD11: DD90 | [308] |
WSM-3978G |
Drug Info | Phase 3 | Hypertrophic cardiomyopathy | ICD11: BC43 | [309] |
ABC294640 |
Drug Info | Phase 3 | Advanced solid tumour | ICD11: 2A00-2F9Z | [310] |
BRN-1907611 |
Drug Info | Phase 3 | Inflammatory bowel disease | ICD11: DD72 | [311] |
DW-1030 |
Drug Info | Phase 3 | Cerebral vasospasm | ICD11: BA85 | [312] |
Fenfluramine |
Drug Info | Phase 3 | Dravet syndrome | ICD11: 8A61-8A6Z | [313] |
Imatinib |
Drug Info | Phase 3 | Mantle cell lymphoma | ICD11: 2A85 | [314] |
ONO-1101 |
Drug Info | Phase 3 | Atrial fibrillation | ICD11: BC81 | [27] |
OPC-12759 |
Drug Info | Phase 3 | Peptic ulcer | ICD11: DA61 | [315] |
RSD-1235 |
Drug Info | Phase 3 | Atrial fibrillation | ICD11: BC81 | [316] |
BNP-1350 |
Drug Info | Phase 3 | Ovarian cancer | ICD11: 2C73 | [317] |
Ganaxolone |
Drug Info | Phase 3 | Complex partial seizure | ICD11: 8A61-8A6Z | [318] |
MRTX849 |
Drug Info | Phase 3 | Lung cancer | ICD11: 2C25 | [319] |
Nitrofural |
Drug Info | Phase 3 | Acute tonsillitis | ICD11: CA03 | [320] |
SEP-363856 |
Drug Info | Phase 3 | Schizophrenia | ICD11: 6A20 | [321] |
Sildenafil |
Drug Info | Phase 3 | Erectile dysfunction | ICD11: HA00-HA01 | [322] |
Ag-221 |
Drug Info | Phase 3 | Acute myelogenous leukaemia | ICD11: 2A41 | [323] |
AMD-070 |
Drug Info | Phase 3 | Whim syndrome | ICD11: 4A00 | [324] |
Elagolix |
Drug Info | Phase 3 | Uterine leiomyoma | ICD11: 2E86 | [325] |
FT-0710230 |
Drug Info | Phase 3 | Opiate dependence | ICD11: 6C43 | [326] |
| Drugs in Phase 2 Clinical Trial | Click to Show/Hide the Full List of Drugs: 42 Drugs | ||||
Emixustat |
Drug Info | Phase 2/3 | Age-related macular degeneration | ICD11: 9B75 | [327] |
Verapamil |
Drug Info | Phase 2/3 | Hypertension | ICD11: BA00-BA04 | [328] |
CS-505 |
Drug Info | Phase 2/3 | Arteriosclerosis obliterans | ICD11: BD40 | [329] |
Fexinidazole |
Drug Info | Phase 2/3 | Trypanosomiasis | ICD11: 1D51-1F53 | [330], [331] |
Fexinidazole |
Drug Info | Phase 2/3 | Trypanosomiasis | ICD11: 1D51-1F53 | [331] |
HOE-239 |
Drug Info | Phase 2/3 | Trypanosomiasis | ICD11: 1F51 | [331] |
MJ-13105 |
Drug Info | Phase 2/3 | Atrial fibrillation | ICD11: BC81 | [27] |
PF-00734200 |
Drug Info | Phase 2 | Type-2 diabetes | ICD11: 5A11 | [332] |
Leniolisib |
Drug Info | Phase 2 | Sjogren syndrome | ICD11: 4A43 | [333] |
AZD8931 |
Drug Info | Phase 2 | Breast cancer | ICD11: 2C60-2C6Y | [334] |
BLZ-945 |
Drug Info | Phase 2 | Amyotrophic lateral sclerosis | ICD11: 8B60 | [335] |
BMS-690514 |
Drug Info | Phase 2 | Chronic pain | ICD11: MG30 | [336] |
BS-3952 |
Drug Info | Phase 2 | Essential hypertension | ICD11: BA00 | [337] |
CCRIS-9277 |
Drug Info | Phase 2 | Posttraumatic stress disorder | ICD11: 6B40 | [338] |
Roxatidine |
Drug Info | Phase 2 | Gastro-oesophageal reflux disease | ICD11: DA22 | [339] |
SQ-109 |
Drug Info | Phase 2 | Pulmonary tuberculosis | ICD11: 1B10 | [340] |
PD-143188 |
Drug Info | Phase 2 | Psychotic disorder | ICD11: 6A20-6A25 | [341] |
PT2385 |
Drug Info | Phase 2 | Von hippel-lindau disease | ICD11: 5A75 | [342] |
VATALANIB |
Drug Info | Phase 2 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [343] |
VATALANIB |
Drug Info | Phase 2 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [344] |
BCP-13498 |
Drug Info | Phase 2 | Anaesthesia | ICD11: 8E22 | [345] |
CP-101,606 |
Drug Info | Phase 2 | Parkinsonism | ICD11: 8A00 | [346] |
SSR-97193 |
Drug Info | Phase 2 | Malaria | ICD11: 1F40 | [347] |
HP-184 |
Drug Info | Phase 2 | Multiple sclerosis | ICD11: 8A40 | [348] |
Org-33062 |
Drug Info | Phase 2 | Cocaine addiction | ICD11: 6C45 | [349] |
Saracatinib |
Drug Info | Phase 2 | Osteosarcoma | ICD11: 2B51 | [350] |
AV-650 |
Drug Info | Phase 2 | Cerebral vasospasm | ICD11: BA85 | [351] |
CP-122721 |
Drug Info | Phase 2 | Depression | ICD11: 6A71 | [352] |
Vincamine |
Drug Info | Phase 2 | Cerebrovascular dementia | ICD11: 6D81 | [353] |
Ym-758 |
Drug Info | Phase 2 | Angina pectoris | ICD11: BA40 | [354] |
JI-101 |
Drug Info | Phase 2 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [355] |
LY-2603618 |
Drug Info | Phase 2 | Pancreatic cancer | ICD11: 2C10 | [356] |
WR-6026 |
Drug Info | Phase 2 | Protozoan infection | ICD11: 1F5Z | [357] |
AZD-2327 |
Drug Info | Phase 2 | Anxiety disorder | ICD11: 6B00-6B0Z | [358] |
CIMICOXIB |
Drug Info | Phase 2 | Pain | ICD11: MG30-MG3Z | [359] |
Nuplazid |
Drug Info | Phase 2 | Alzheimer disease | ICD11: 8A20 | [360] |
PTC299 |
Drug Info | Phase 2 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [361] |
TAK-906 |
Drug Info | Phase 2 | Gastroparesis | ICD11: DA41 | [362] |
KD025 |
Drug Info | Phase 2 | Psoriasis vulgaris | ICD11: EA90 | [363] |
KW-4354 |
Drug Info | Phase 2 | Muscular dystrophy | ICD11: 8C70 | [364] |
P-459 |
Drug Info | Phase 2 | Staphylococcus infection | ICD11: 1B73 | [55] |
Prajmaline |
Drug Info | Phase 2 | Atherosclerosis | ICD11: BA80 | [365] |
| Drugs in Phase 1 Clinical Trial | Click to Show/Hide the Full List of Drugs: 16 Drugs | ||||
G-33040 |
Drug Info | Phase 1/2 | Anxiety disorder | ICD11: 6B00 | [55] |
TRV-130 |
Drug Info | Phase 1/2 | Hypothyroidism | ICD11: 5A00 | [366] |
GTPL7557 |
Drug Info | Phase 1/2 | Schizophrenia | ICD11: 6A20 | [367] |
AST-1306 |
Drug Info | Phase 1 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [368] |
BE-14348A |
Drug Info | Phase 1 | Viral hepatitis | ICD11: 1E51 | [369] |
H3B-6545 |
Drug Info | Phase 1 | Breast cancer | ICD11: 2C60 | [370] |
M-813 |
Drug Info | Phase 1 | Schizophrenia | ICD11: 6A20 | [371] |
Alvespimycin hydrochloride |
Drug Info | Phase 1 | Ovarian cancer | ICD11: 2C73 | [372] |
Benzoylphenylurea |
Drug Info | Phase 1 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [373], [374] |
MP-513 |
Drug Info | Phase 1 | Diabetes mellitus | ICD11: 5A10 | [375], [376] |
AC-1802 |
Drug Info | Phase 1 | Cardiac arrhythmia | ICD11: BC65 | [377] |
CCRIS-4201 |
Drug Info | Phase 1 | Essential hypertension | ICD11: BA00 | [27] |
GTPL-1666 |
Drug Info | Phase 1 | Cerebral vasospasm | ICD11: BA85 | [378] |
NSC-172130 |
Drug Info | Phase 1 | Depression | ICD11: 6A71 | [379] |
OSI-930 |
Drug Info | Phase 1 | Solid tumour/cancer | ICD11: 2A00-2F9Z | [380] |
ITX-5061 |
Drug Info | Phase 1 | Human immunodeficiency virus infection | ICD11: 1C60 | [381] |
| Discontinued/withdrawn Drugs | Click to Show/Hide the Full List of Drugs: 24 Drugs | ||||
Cilomilast |
Drug Info | Discontinued in Phase 3 | Emphysema | ICD11: CA21 | [382] |
Motesanib |
Drug Info | Discontinued in Phase 3 | Lung cancer | ICD11: 2C25 | [383] |
Almokalant |
Drug Info | Discontinued in Phase 3 | Cardiac arrhythmias | ICD11: BC9Z | [384] |
Avitriptan |
Drug Info | Discontinued in Phase 3 | Migraine | ICD11: 8A80 | [385] |
AZD0328 |
Drug Info | Discontinued in Phase 2 | Alzheimer disease | ICD11: 8A20 | [386] |
TAK-802 |
Drug Info | Discontinued in Phase 2 | Urinary dysfunction | ICD11: GC2Z | [387] |
YM-992 |
Drug Info | Discontinued in Phase 2 | Depression | ICD11: 6A70-6A7Z | [388] |
BMS-181168 |
Drug Info | Discontinued in Phase 2 | Cognitive impairment | ICD11: 6D71 | [389] |
Ispronicline |
Drug Info | Discontinued in Phase 2 | Schizophrenia | ICD11: 6A20 | [390] |
ABT-107 |
Drug Info | Discontinued in Phase 1 | Attention deficit hyperactivity disorder | ICD11: 6A05 | [391] |
E2101 |
Drug Info | Discontinued in Phase 1 | Dyskinesia | ICD11: 8A02 | [392] |
YM-17E |
Drug Info | Discontinued in Phase 1 | Hypertriglyceridaemia | ICD11: 5C80 | [393] |
CJ-11974 |
Drug Info | Discontinued | Irritable bowel syndrome | ICD11: DD91 | [394] |
CAM-2028 |
Drug Info | Discontinued | Acute nasopharyngitis | ICD11: CA00 | [395] |
ML-3000 |
Drug Info | Discontinued | Osteoarthritis | ICD11: FA00 | [13] |
BMY-42569 |
Drug Info | Discontinued | Depression | ICD11: 6A71 | [396] |
R-58735 |
Drug Info | Discontinued | Cerebral ischaemia | ICD11: 8B1Z | [397] |
CGP-11305A |
Drug Info | Discontinued | Anxiety disorder | ICD11: 6B00 | [398] |
INN-00835 |
Drug Info | Discontinued | Depression | ICD11: 6A71 | [399] |
Octopamine |
Drug Info | Discontinued | Thrombosis | ICD11: DB61 | [400] |
RD-4593 |
Drug Info | Discontinued | Depression | ICD11: 6A71 | [401] |
TS-702 |
Drug Info | Discontinued | Angina pectoris | ICD11: BA40 | [27] |
TV-1203 |
Drug Info | Discontinued | Parkinsonism | ICD11: 8A00 | [402] |
YMB-1002 |
Drug Info | Discontinued | Breast cancer | ICD11: 2C60 | [403] |
| Preclinical/investigative Agents | Click to Show/Hide the Full List of Drugs: 23 Drugs | ||||
EPX-100 |
Drug Info | Preclinical | Dravet syndrome | ICD11: 8A61 | [404] |
Q-100155 |
Drug Info | Preclinical | Rheumatoid arthritis | ICD11: FA20 | [405] |
Befunolol |
Drug Info | Investigative | Glaucoma | ICD11: 9C61 | [27] |
Ethylmorphine |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [406] |
Bufuralol |
Drug Info | Investigative | Cardiac arrhythmia | ICD11: BC65 | [407] |
Butyrfentanyl |
Drug Info | Investigative | Anaesthesia | ICD11: 8E22 | [408] |
Bromazepam |
Drug Info | Investigative | Anxiety disorder | ICD11: 6B00 | [409] |
Methoxyamphetamine beta |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [410] |
Antipyrine |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [411] |
Chloroxylenol |
Drug Info | Investigative | Pseudomonas infection | ICD11: 1G40 | [55] |
Desmethylastemizole |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [265] |
Linalool |
Drug Info | Investigative | Colon cancer | ICD11: 2B90 | [412], [413], [414] |
Methoxyphenamine |
Drug Info | Investigative | Chronic obstructive pulmonary disease | ICD11: CA22 | [415] |
Bupranolol |
Drug Info | Investigative | Ventricular tachyarrhythmia | ICD11: BC71 | [55] |
Mequitazine |
Drug Info | Investigative | Allergic rhinitis | ICD11: CA08 | [416] |
Methoxybufotenin |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [417] |
Sparteine |
Drug Info | Investigative | Cardiac arrhythmia | ICD11: BC65 | [131] |
Bopindolol |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [27] |
Etilamfetamine |
Drug Info | Investigative | Pulmonary hypertension | ICD11: BB01 | [157] |
Lorpiprazole |
Drug Info | Investigative | Anxiety disorder | ICD11: 6B00 | [418] |
Repinotan |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [419] |
Cloranolol |
Drug Info | Investigative | Discovery agent | ICD: N.A. | [27] |
Mepindolol |
Drug Info | Investigative | Glaucoma | ICD11: 9C61 | [27] |
| Tissue/Disease-Specific Protein Abundances of This DME | |||||
|---|---|---|---|---|---|
| Tissue-specific Protein Abundances in Healthy Individuals | Click to Show/Hide | ||||
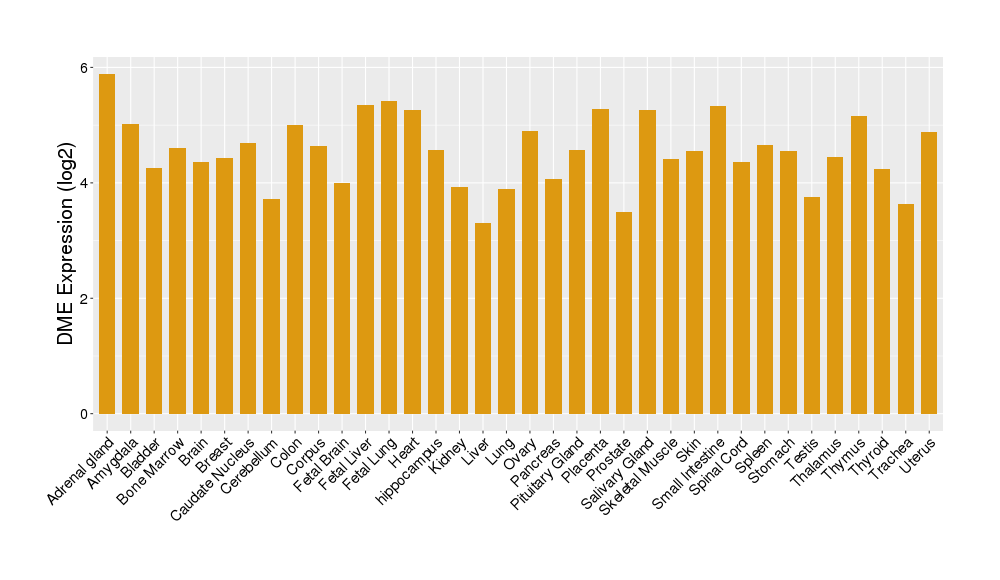
|
|||||
| ICD Disease Classification 01 | Infectious/parasitic disease | Click to Show/Hide | |||
| ICD-11: 1C1H | Necrotising ulcerative gingivitis | Click to Show/Hide | |||
| The Studied Tissue | Gingival tissue | ||||
| The Specified Disease | Bacterial infection of gingival [ICD-11:1C1H] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.83E-01; Fold-change: -1.74E-02; Z-score: -7.78E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
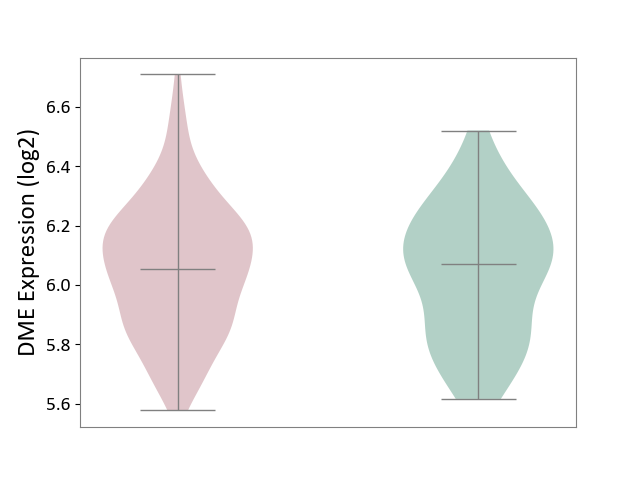
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 1E30 | Influenza | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Influenza [ICD-11:1E30] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.29E-04; Fold-change: 4.43E-01; Z-score: 1.04E+01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
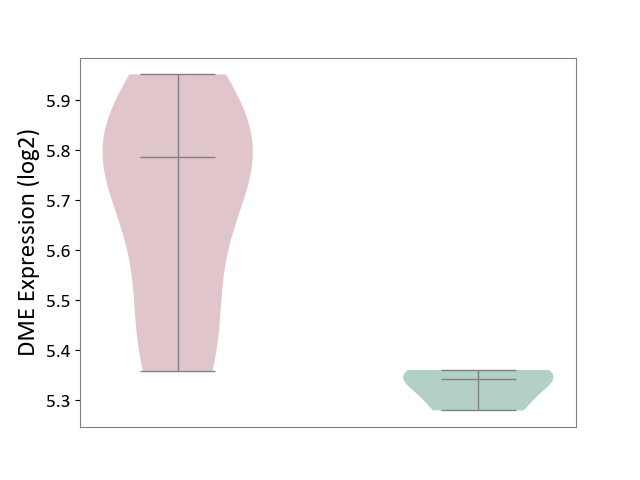
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 1E51 | Chronic viral hepatitis | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Chronic hepatitis C [ICD-11:1E51.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.31E-01; Fold-change: 1.04E-01; Z-score: 8.98E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
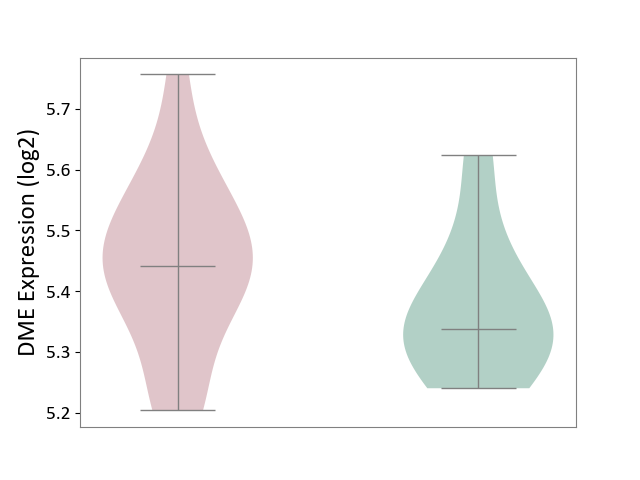
|
Click to View the Clearer Original Diagram | |||
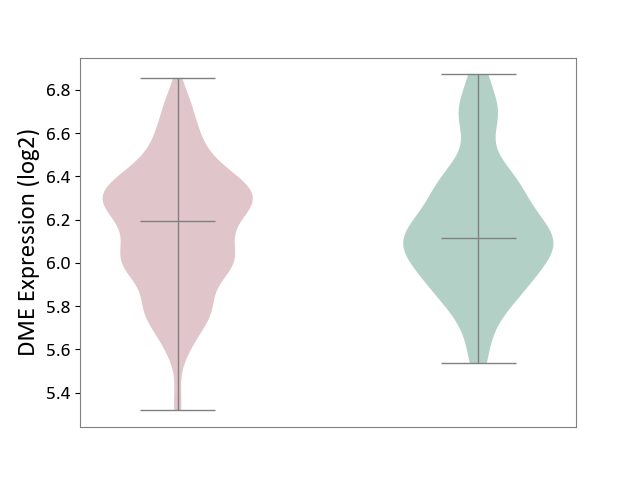
| ICD-11: 1G41 | Sepsis with septic shock | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Sepsis with septic shock [ICD-11:1G41] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.79E-01; Fold-change: 7.87E-02; Z-score: 2.63E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
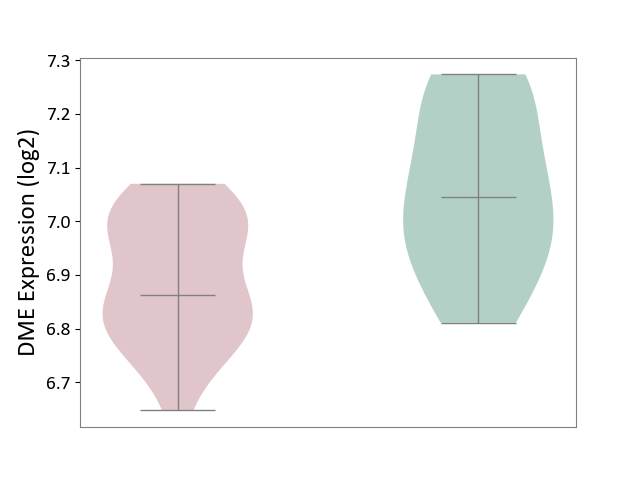
| ICD-11: CA40 | Respiratory syncytial virus infection | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Pediatric respiratory syncytial virus infection [ICD-11:CA40.11] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.24E-04; Fold-change: -1.83E-01; Z-score: -1.21E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
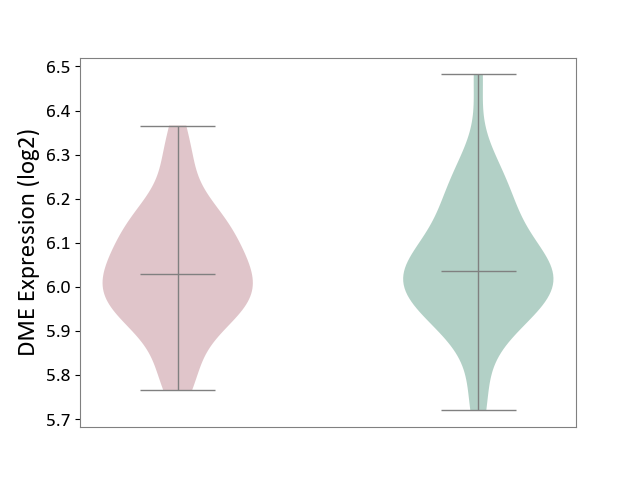
| ICD-11: CA42 | Rhinovirus infection | Click to Show/Hide | |||
| The Studied Tissue | Nasal epithelium tissue | ||||
| The Specified Disease | Rhinovirus infection [ICD-11:CA42.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.00E-01; Fold-change: -7.71E-03; Z-score: -5.19E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
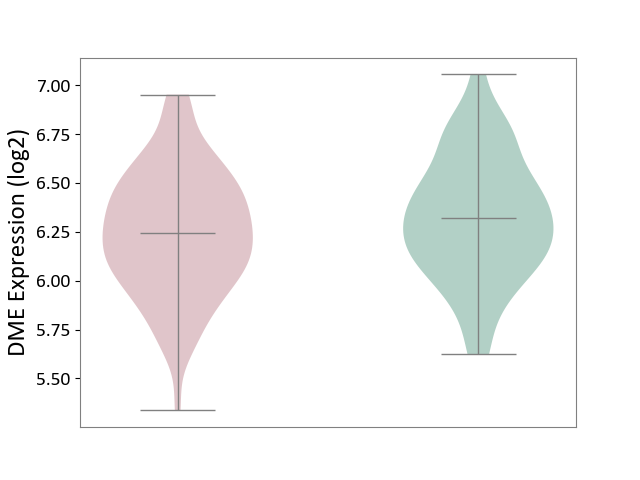
| ICD-11: KA60 | Neonatal sepsis | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Neonatal sepsis [ICD-11:KA60] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.68E-02; Fold-change: -7.62E-02; Z-score: -2.41E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 02 | Neoplasm | Click to Show/Hide | |||
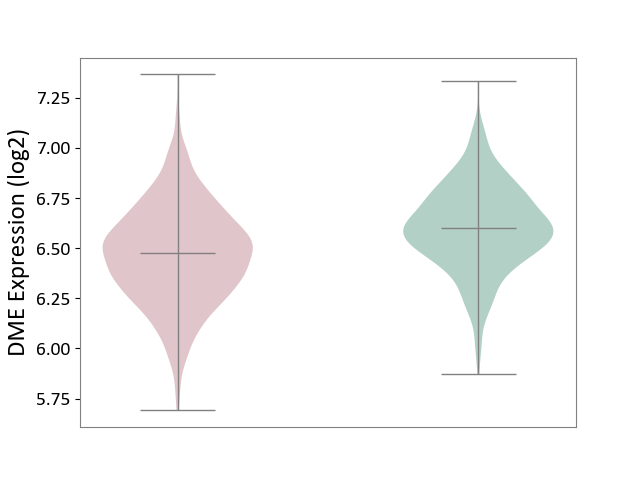
| ICD-11: 2A00 | Brain cancer | Click to Show/Hide | |||
| The Studied Tissue | Nervous tissue | ||||
| The Specified Disease | Glioblastopma [ICD-11:2A00.00] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.16E-22; Fold-change: -1.26E-01; Z-score: -5.93E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
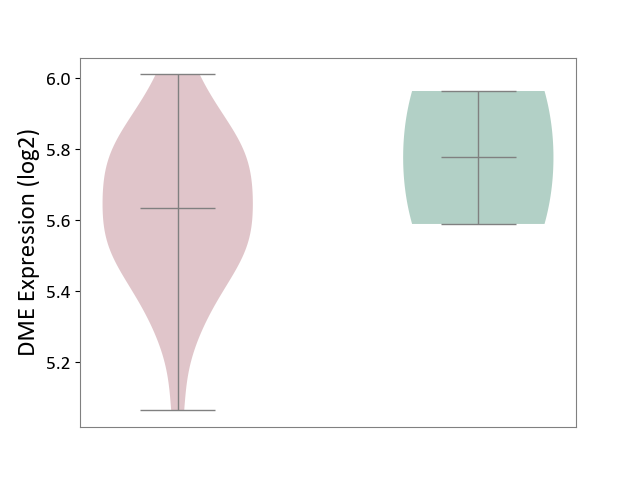
| The Studied Tissue | Brain stem tissue | ||||
| The Specified Disease | Glioma [ICD-11:2A00.0Y-2A00.0Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.73E-01; Fold-change: -1.42E-01; Z-score: -5.35E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
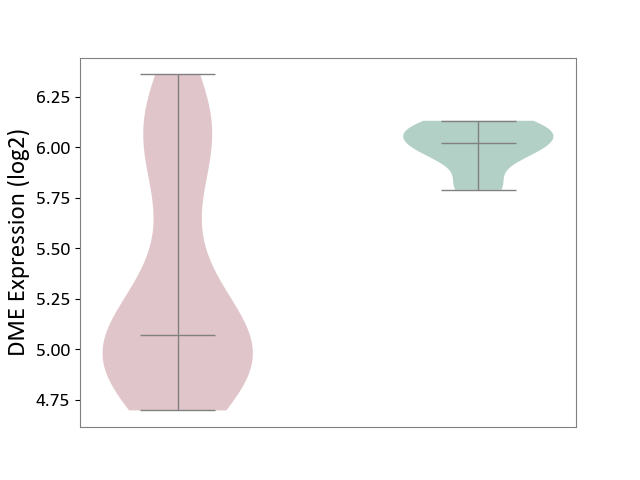
| The Studied Tissue | White matter tissue | ||||
| The Specified Disease | Glioma [ICD-11:2A00.0Y-2A00.0Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.09E-09; Fold-change: -9.49E-01; Z-score: -8.32E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
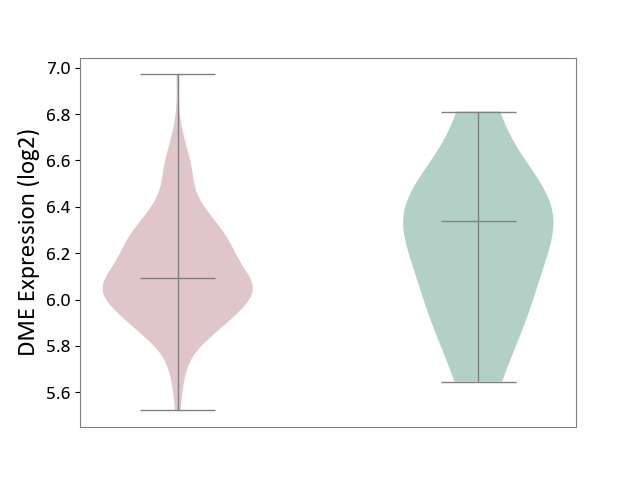
| The Studied Tissue | Brain stem tissue | ||||
| The Specified Disease | Neuroectodermal tumour [ICD-11:2A00.11] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.34E-01; Fold-change: -2.49E-01; Z-score: -7.53E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2A20 | Myeloproliferative neoplasm | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Myelofibrosis [ICD-11:2A20.2] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.41E-01; Fold-change: -1.92E-02; Z-score: -2.46E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
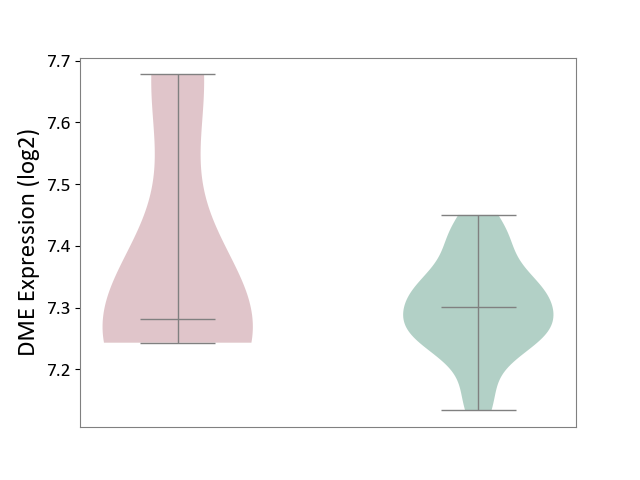
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Polycythemia vera [ICD-11:2A20.4] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.22E-02; Fold-change: -2.51E-02; Z-score: -2.94E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
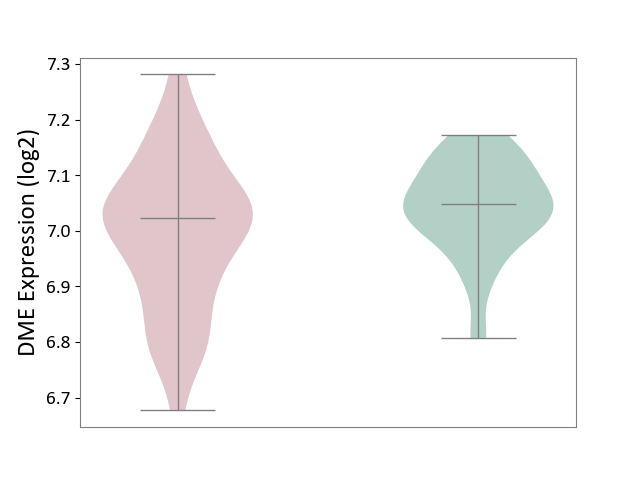
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2A36 | Myelodysplastic syndrome | Click to Show/Hide | |||
| The Studied Tissue | Bone marrow | ||||
| The Specified Disease | Myelodysplastic syndrome [ICD-11:2A36-2A3Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.26E-01; Fold-change: 5.09E-02; Z-score: 2.01E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.93E-03; Fold-change: -4.65E-01; Z-score: -4.51E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
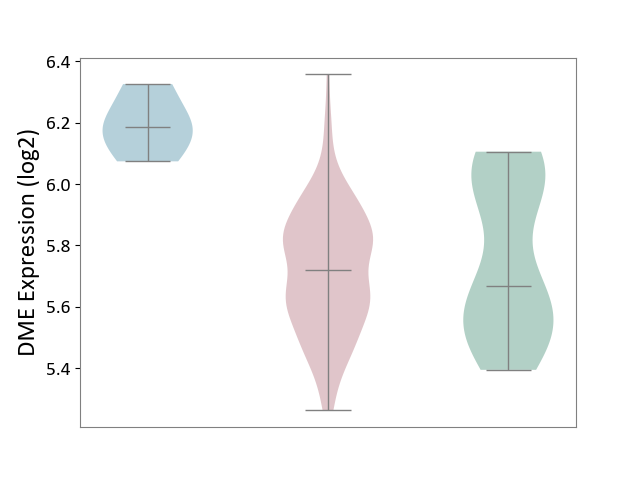
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2A81 | Diffuse large B-cell lymphoma | Click to Show/Hide | |||
| The Studied Tissue | Tonsil tissue | ||||
| The Specified Disease | Diffuse large B-cell lymphoma [ICD-11:2A81] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.88E-01; Fold-change: -1.28E-01; Z-score: -5.96E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
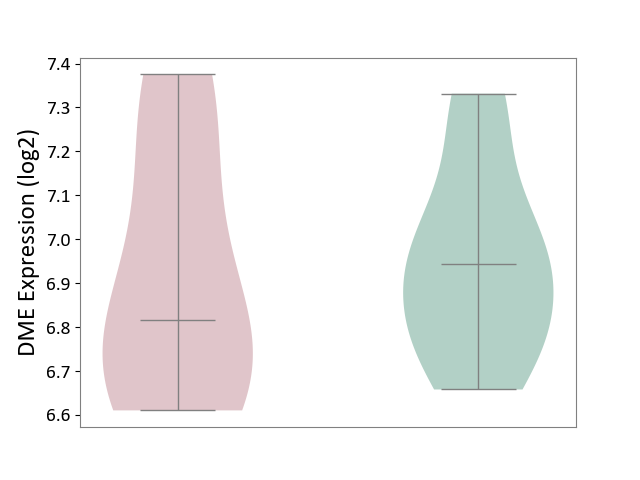
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2A83 | Plasma cell neoplasm | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Multiple myeloma [ICD-11:2A83.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.28E-01; Fold-change: -3.69E-02; Z-score: -1.61E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
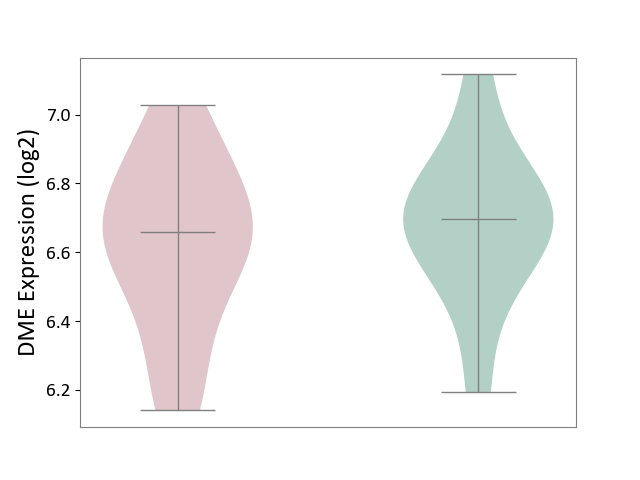
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Bone marrow | ||||
| The Specified Disease | Multiple myeloma [ICD-11:2A83.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.35E-01; Fold-change: -5.39E-03; Z-score: -5.20E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
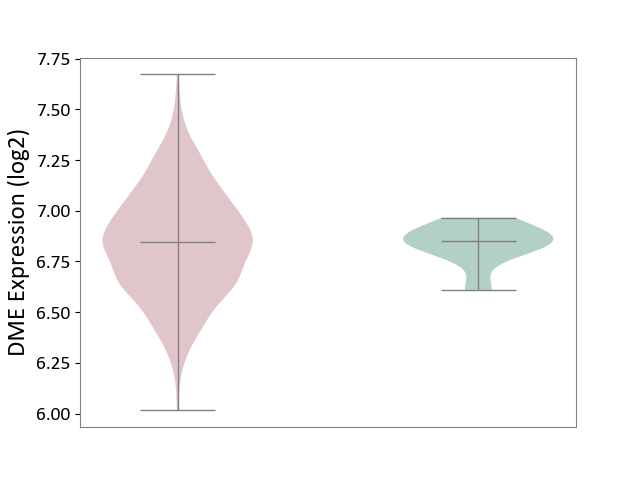
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2B33 | Leukaemia | Click to Show/Hide | |||
| The Studied Tissue | Bone marrow | ||||
| The Specified Disease | Acute myelocytic leukaemia [ICD-11:2B33.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.68E-01; Fold-change: 5.03E-03; Z-score: 2.52E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
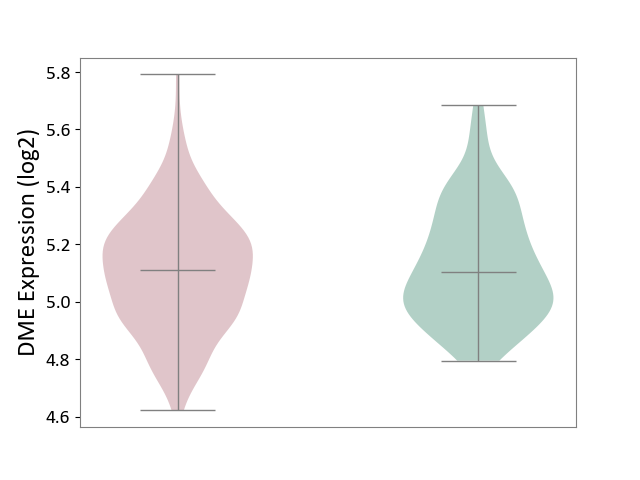
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2B6E | Oral cancer | Click to Show/Hide | |||
| The Studied Tissue | Oral tissue | ||||
| The Specified Disease | Oral cancer [ICD-11:2B6E] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.10E-11; Fold-change: -3.94E-01; Z-score: -1.97E+00 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.89E-01; Fold-change: 2.68E-02; Z-score: 1.03E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
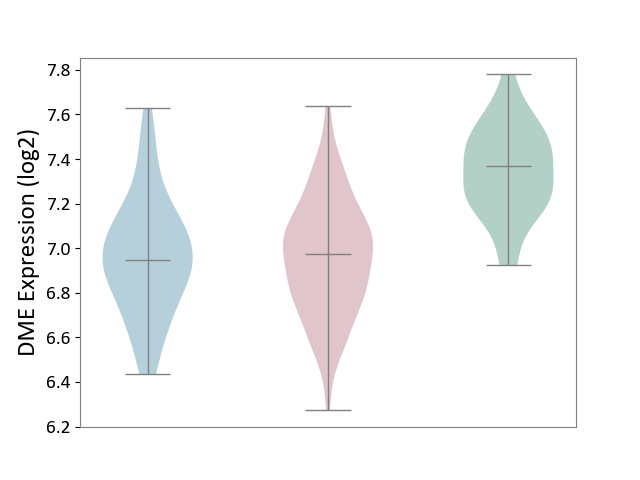
|
Click to View the Clearer Original Diagram | |||
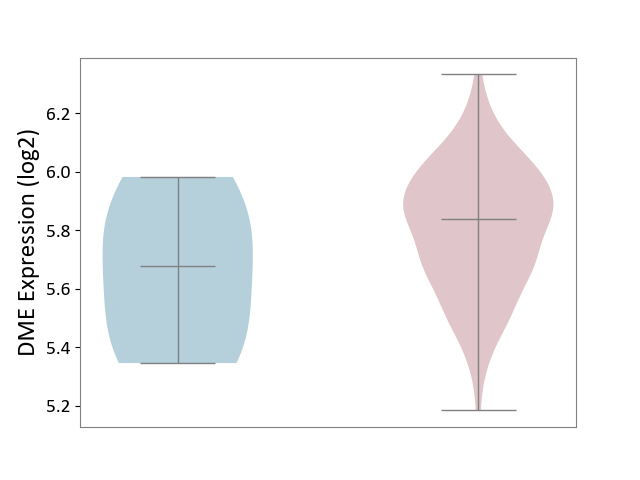
| ICD-11: 2B70 | Esophageal cancer | Click to Show/Hide | |||
| The Studied Tissue | Esophagus | ||||
| The Specified Disease | Esophagal cancer [ICD-11:2B70] | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.63E-01; Fold-change: 1.59E-01; Z-score: 6.06E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
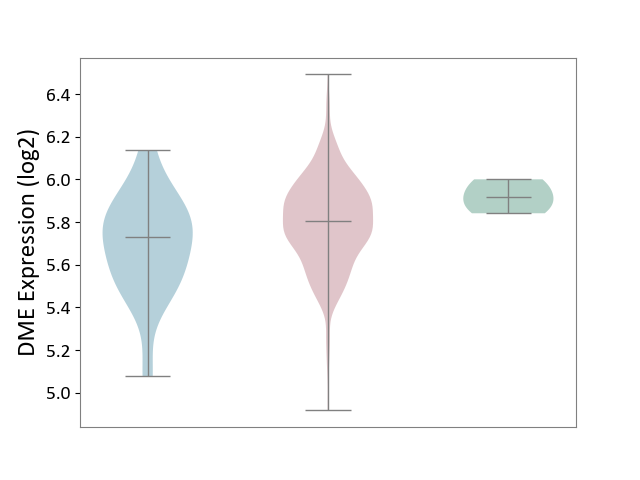
| ICD-11: 2B72 | Stomach cancer | Click to Show/Hide | |||
| The Studied Tissue | Gastric tissue | ||||
| The Specified Disease | Gastric cancer [ICD-11:2B72] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.16E-01; Fold-change: -1.10E-01; Z-score: -1.38E+00 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.45E-02; Fold-change: 7.74E-02; Z-score: 3.22E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
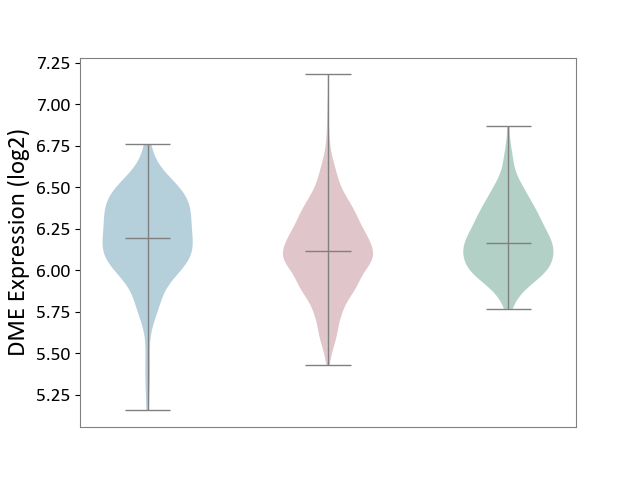
| ICD-11: 2B90 | Colon cancer | Click to Show/Hide | |||
| The Studied Tissue | Colon tissue | ||||
| The Specified Disease | Colon cancer [ICD-11:2B90] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.39E-05; Fold-change: -5.40E-02; Z-score: -2.65E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.13E-03; Fold-change: -8.16E-02; Z-score: -3.02E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
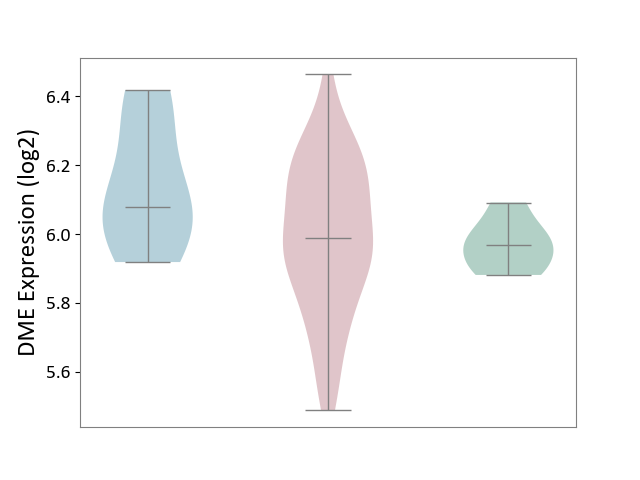
| ICD-11: 2B92 | Rectal cancer | Click to Show/Hide | |||
| The Studied Tissue | Rectal colon tissue | ||||
| The Specified Disease | Rectal cancer [ICD-11:2B92] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.98E-01; Fold-change: 1.97E-02; Z-score: 2.59E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.19E-01; Fold-change: -9.12E-02; Z-score: -4.83E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
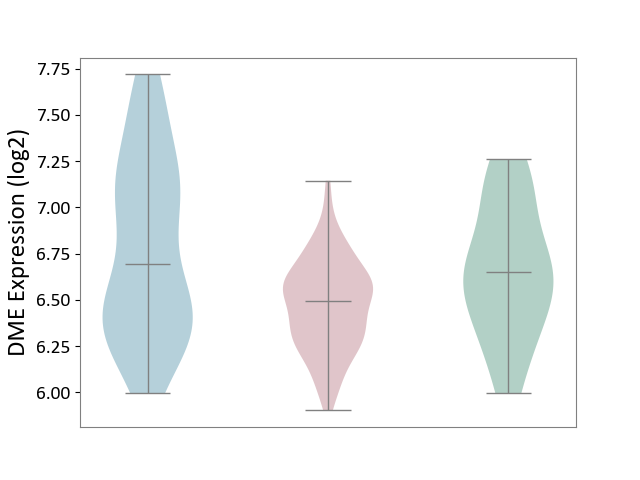
| ICD-11: 2C10 | Pancreatic cancer | Click to Show/Hide | |||
| The Studied Tissue | Pancreas | ||||
| The Specified Disease | Pancreatic cancer [ICD-11:2C10] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.44E-02; Fold-change: -1.56E-01; Z-score: -4.37E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.57E-05; Fold-change: -1.97E-01; Z-score: -4.11E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
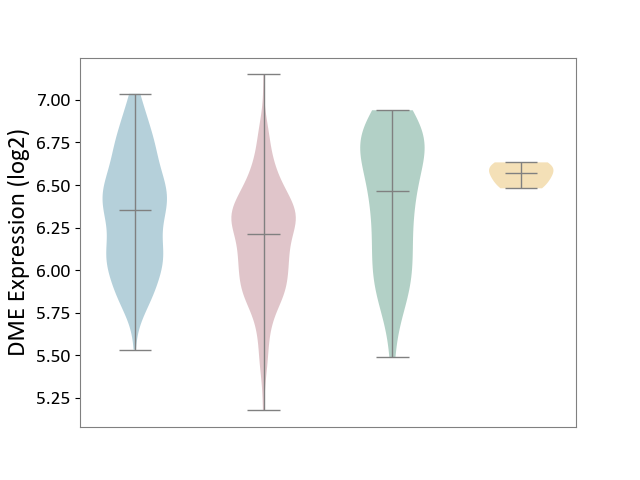
| ICD-11: 2C12 | Liver cancer | Click to Show/Hide | |||
| The Studied Tissue | Liver tissue | ||||
| The Specified Disease | Liver cancer [ICD-11:2C12.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.63E-04; Fold-change: -2.52E-01; Z-score: -6.62E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.24E-08; Fold-change: -1.43E-01; Z-score: -4.34E-01 | ||||
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 2.26E-04; Fold-change: -3.58E-01; Z-score: -5.37E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
DME expression in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
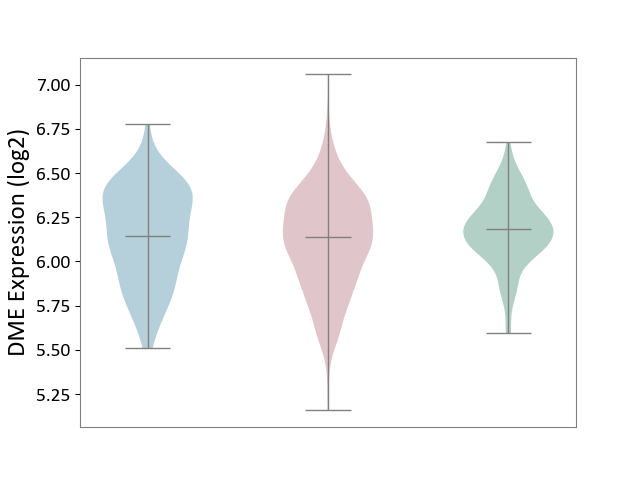
| ICD-11: 2C25 | Lung cancer | Click to Show/Hide | |||
| The Studied Tissue | Lung tissue | ||||
| The Specified Disease | Lung cancer [ICD-11:2C25] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.76E-04; Fold-change: -4.73E-02; Z-score: -2.39E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.38E-01; Fold-change: -9.66E-03; Z-score: -3.59E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
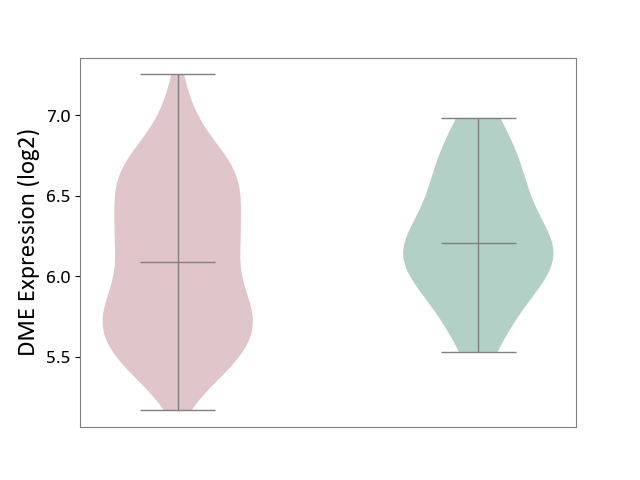
| ICD-11: 2C30 | Skin cancer | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Melanoma [ICD-11:2C30] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.64E-02; Fold-change: -1.20E-01; Z-score: -3.11E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
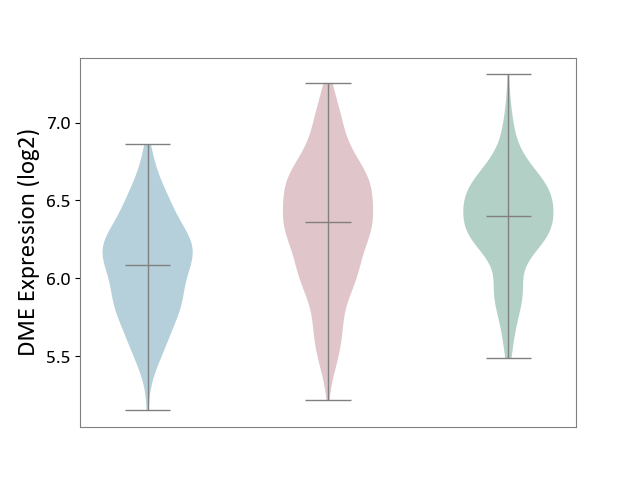
| The Studied Tissue | Skin | ||||
| The Specified Disease | Skin cancer [ICD-11:2C30-2C3Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.37E-01; Fold-change: -3.80E-02; Z-score: -1.16E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.00E-15; Fold-change: 2.76E-01; Z-score: 8.36E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
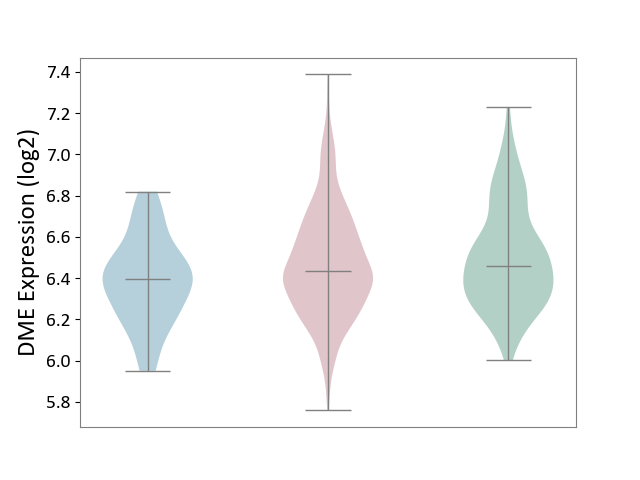
| ICD-11: 2C6Z | Breast cancer | Click to Show/Hide | |||
| The Studied Tissue | Breast tissue | ||||
| The Specified Disease | Breast cancer [ICD-11:2C60-2C6Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.51E-02; Fold-change: -2.38E-02; Z-score: -9.58E-02 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.14E-02; Fold-change: 4.23E-02; Z-score: 2.03E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
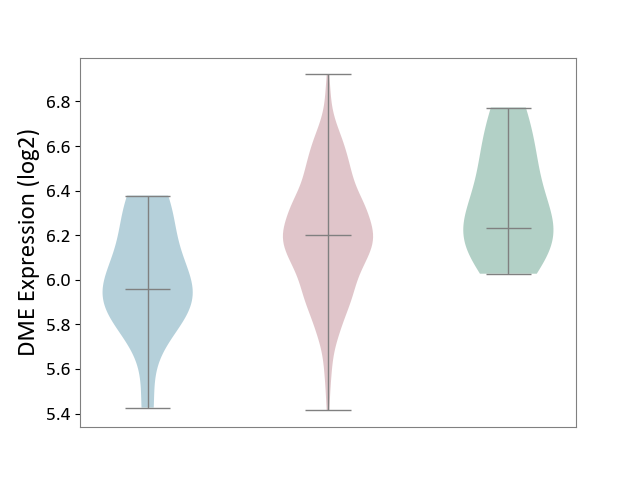
| ICD-11: 2C73 | Ovarian cancer | Click to Show/Hide | |||
| The Studied Tissue | Ovarian tissue | ||||
| The Specified Disease | Ovarian cancer [ICD-11:2C73] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.02E-01; Fold-change: -3.26E-02; Z-score: -1.33E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.28E-02; Fold-change: 2.40E-01; Z-score: 9.51E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
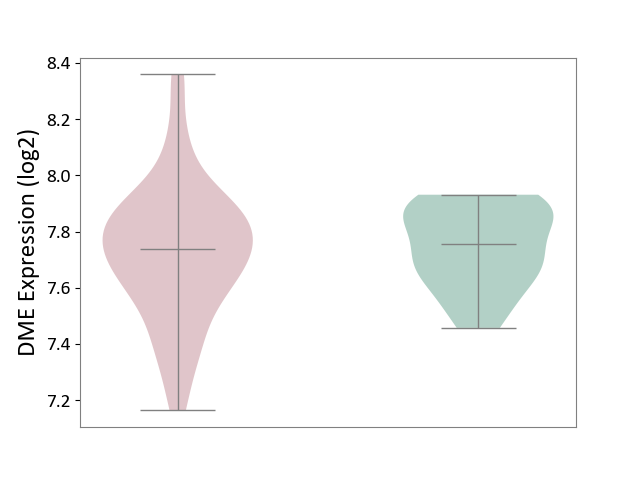
| ICD-11: 2C77 | Cervical cancer | Click to Show/Hide | |||
| The Studied Tissue | Cervical tissue | ||||
| The Specified Disease | Cervical cancer [ICD-11:2C77] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.95E-01; Fold-change: -2.05E-02; Z-score: -1.41E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C78 | Uterine cancer | Click to Show/Hide | |||
| The Studied Tissue | Endometrium tissue | ||||
| The Specified Disease | Uterine cancer [ICD-11:2C78] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.27E-02; Fold-change: -6.33E-02; Z-score: -2.52E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.17E-03; Fold-change: -9.69E-02; Z-score: -1.70E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
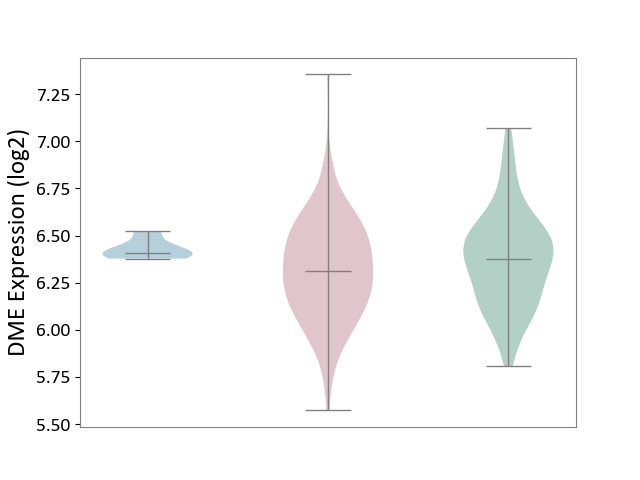
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C82 | Prostate cancer | Click to Show/Hide | |||
| The Studied Tissue | Prostate | ||||
| The Specified Disease | Prostate cancer [ICD-11:2C82] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.66E-01; Fold-change: -1.19E-01; Z-score: -3.65E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
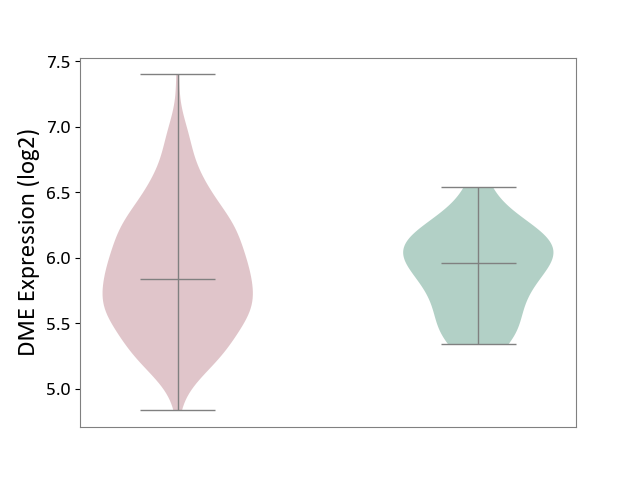
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C90 | Renal cancer | Click to Show/Hide | |||
| The Studied Tissue | Kidney | ||||
| The Specified Disease | Renal cancer [ICD-11:2C90-2C91] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.59E-01; Fold-change: -4.51E-02; Z-score: -1.36E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.16E-11; Fold-change: -1.98E-01; Z-score: -9.26E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
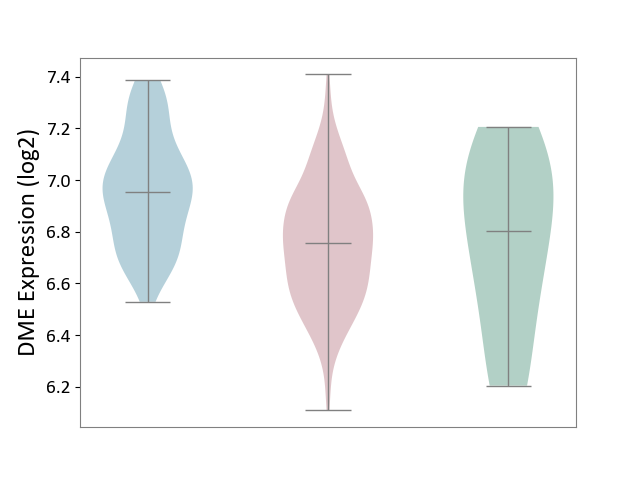
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C92 | Ureter cancer | Click to Show/Hide | |||
| The Studied Tissue | Urothelium | ||||
| The Specified Disease | Ureter cancer [ICD-11:2C92] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.68E-01; Fold-change: -3.39E-02; Z-score: -1.30E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
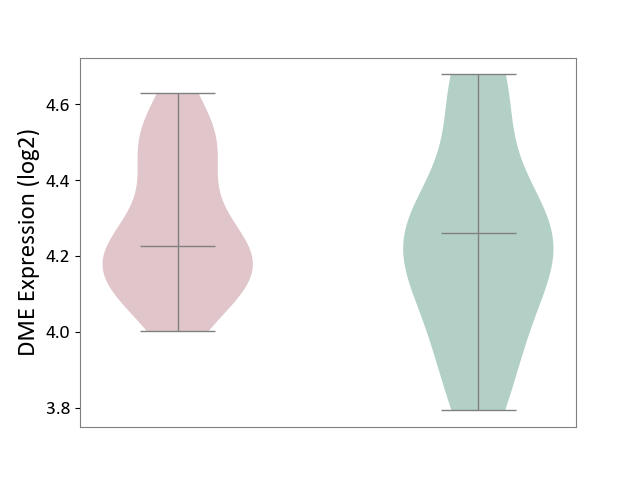
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2C94 | Bladder cancer | Click to Show/Hide | |||
| The Studied Tissue | Bladder tissue | ||||
| The Specified Disease | Bladder cancer [ICD-11:2C94] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.53E-04; Fold-change: 7.65E-01; Z-score: 2.65E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
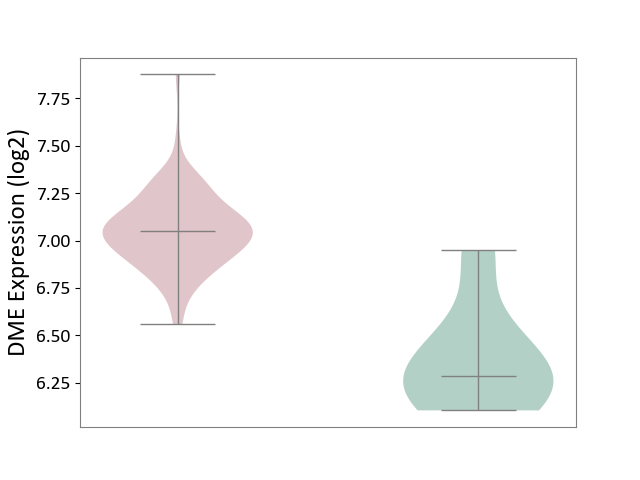
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2D02 | Retinal cancer | Click to Show/Hide | |||
| The Studied Tissue | Uvea | ||||
| The Specified Disease | Retinoblastoma [ICD-11:2D02.2] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.21E-06; Fold-change: -4.93E-01; Z-score: -6.09E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
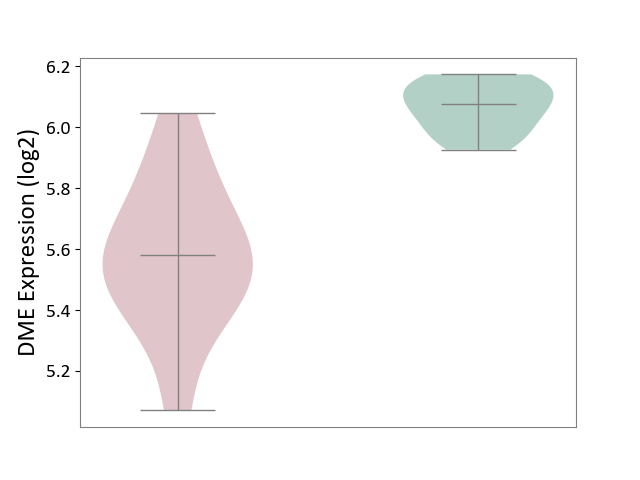
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2D10 | Thyroid cancer | Click to Show/Hide | |||
| The Studied Tissue | Thyroid | ||||
| The Specified Disease | Thyroid cancer [ICD-11:2D10] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.46E-07; Fold-change: 2.36E-01; Z-score: 7.25E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.64E-02; Fold-change: 3.70E-02; Z-score: 1.52E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
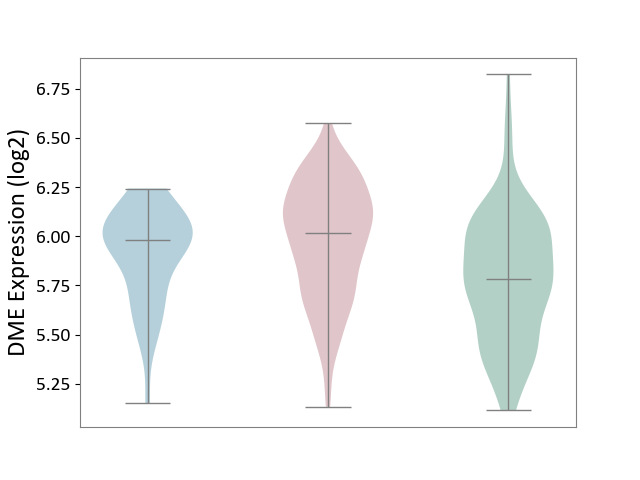
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 2D11 | Adrenal cancer | Click to Show/Hide | |||
| The Studied Tissue | Adrenal cortex | ||||
| The Specified Disease | Adrenocortical carcinoma [ICD-11:2D11.Z] | ||||
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 5.95E-02; Fold-change: -1.76E-01; Z-score: -1.04E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
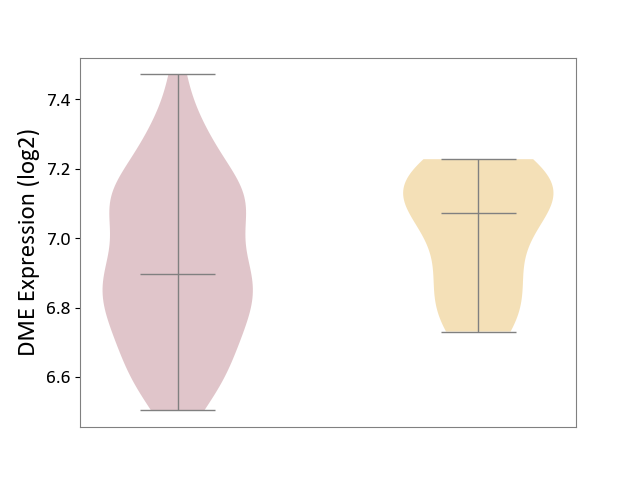
|
Click to View the Clearer Original Diagram | |||
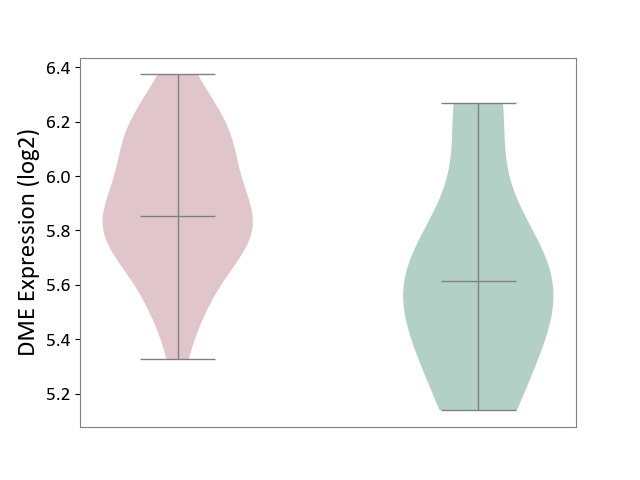
| ICD-11: 2D12 | Endocrine gland neoplasm | Click to Show/Hide | |||
| The Studied Tissue | Pituitary tissue | ||||
| The Specified Disease | Pituitary cancer [ICD-11:2D12] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.79E-02; Fold-change: 2.40E-01; Z-score: 6.54E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
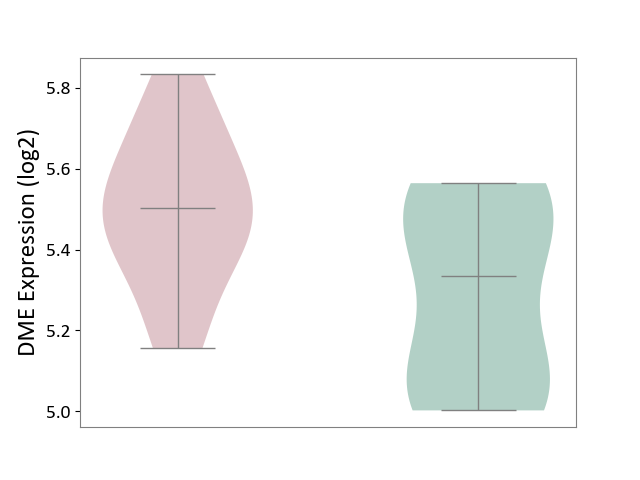
| The Studied Tissue | Pituitary tissue | ||||
| The Specified Disease | Pituitary gonadotrope tumour [ICD-11:2D12] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.16E-02; Fold-change: 1.69E-01; Z-score: 7.14E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
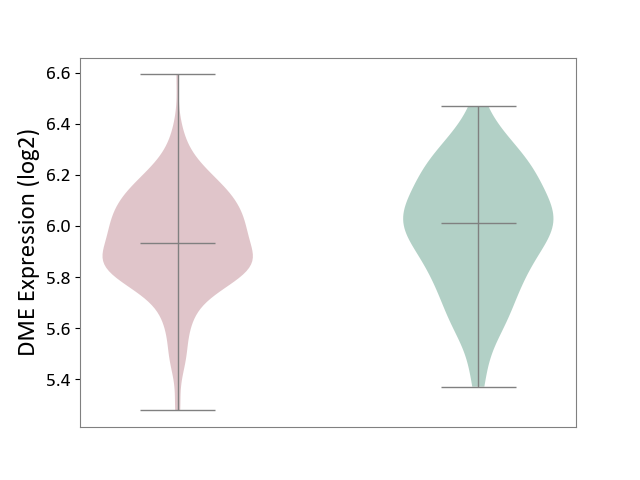
| ICD-11: 2D42 | Head and neck cancer | Click to Show/Hide | |||
| The Studied Tissue | Head and neck tissue | ||||
| The Specified Disease | Head and neck cancer [ICD-11:2D42] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.14E-01; Fold-change: -7.61E-02; Z-score: -3.24E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 03 | Blood/blood-forming organ disease | Click to Show/Hide | |||
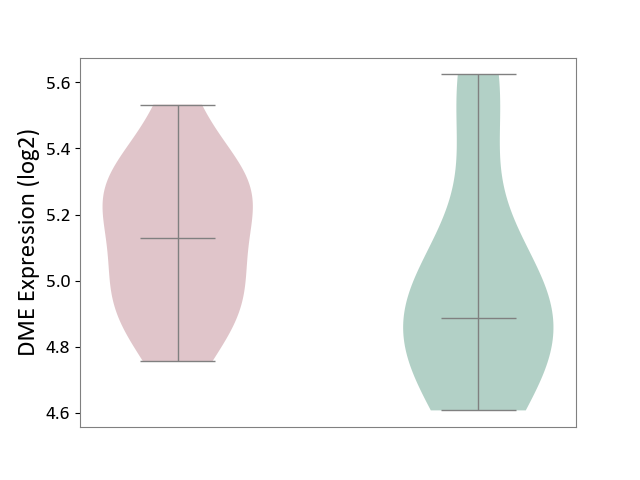
| ICD-11: 3A51 | Sickle cell disorder | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Sickle cell disease [ICD-11:3A51.0-3A51.3] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.28E-01; Fold-change: 2.43E-01; Z-score: 7.45E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
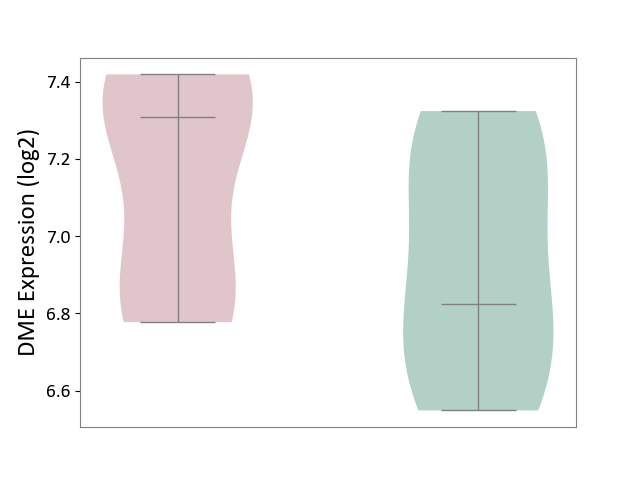
| ICD-11: 3A70 | Aplastic anaemia | Click to Show/Hide | |||
| The Studied Tissue | Bone marrow | ||||
| The Specified Disease | Shwachman-Diamond syndrome [ICD-11:3A70.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.13E-01; Fold-change: 4.85E-01; Z-score: 1.58E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
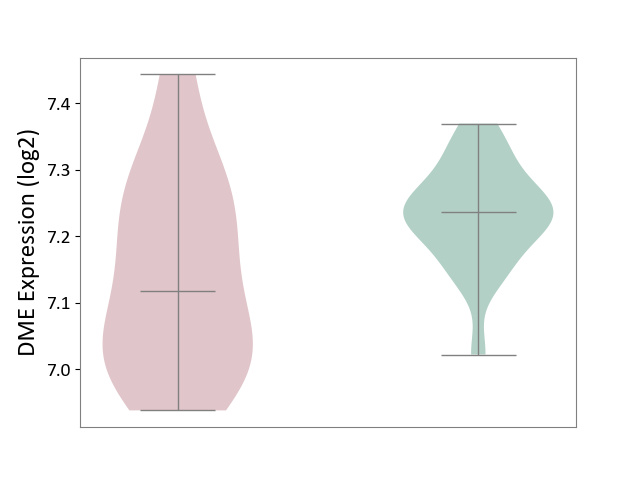
| ICD-11: 3B63 | Thrombocytosis | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Thrombocythemia [ICD-11:3B63] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.76E-03; Fold-change: -1.19E-01; Z-score: -1.54E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
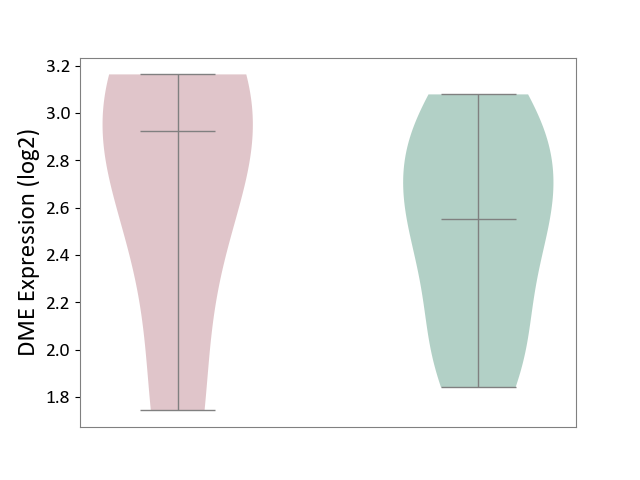
| ICD-11: 3B64 | Thrombocytopenia | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Thrombocytopenia [ICD-11:3B64] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.86E-01; Fold-change: 3.72E-01; Z-score: 8.87E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 04 | Immune system disease | Click to Show/Hide | |||
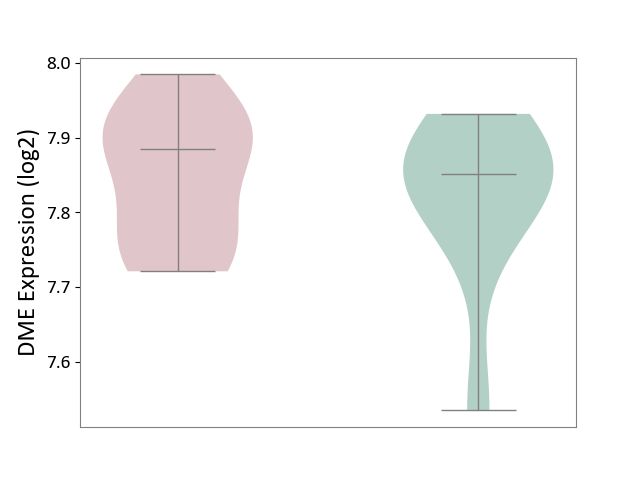
| ICD-11: 4A00 | Immunodeficiency | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Immunodeficiency [ICD-11:4A00-4A20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.53E-01; Fold-change: 3.38E-02; Z-score: 2.92E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 4A40 | Lupus erythematosus | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Lupus erythematosus [ICD-11:4A40] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.24E-03; Fold-change: -3.04E-01; Z-score: -6.23E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
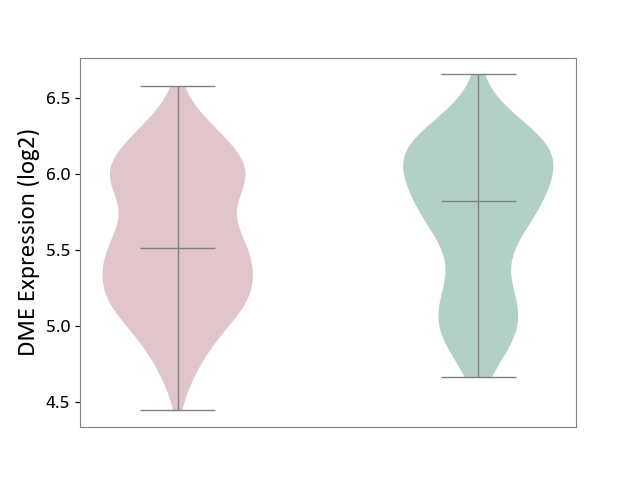
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 4A42 | Systemic sclerosis | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Scleroderma [ICD-11:4A42.Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.28E-02; Fold-change: 8.85E-02; Z-score: 8.03E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
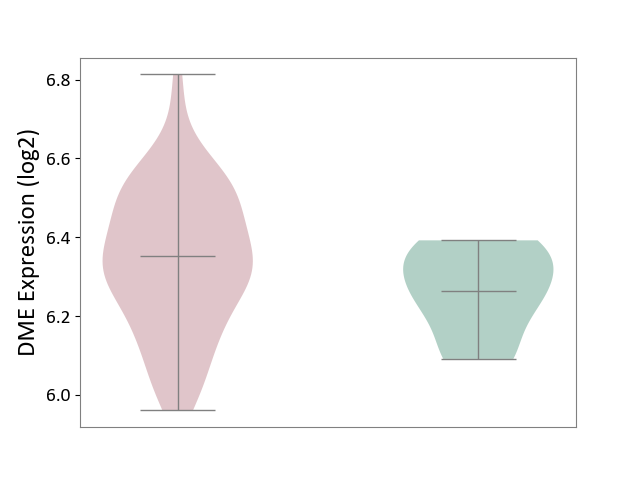
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 4A43 | Systemic autoimmune disease | Click to Show/Hide | |||
| The Studied Tissue | Salivary gland tissue | ||||
| The Specified Disease | Sjogren's syndrome [ICD-11:4A43.2] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.44E-02; Fold-change: -5.80E-01; Z-score: -1.74E+00 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.78E-01; Fold-change: 1.62E-01; Z-score: 7.72E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
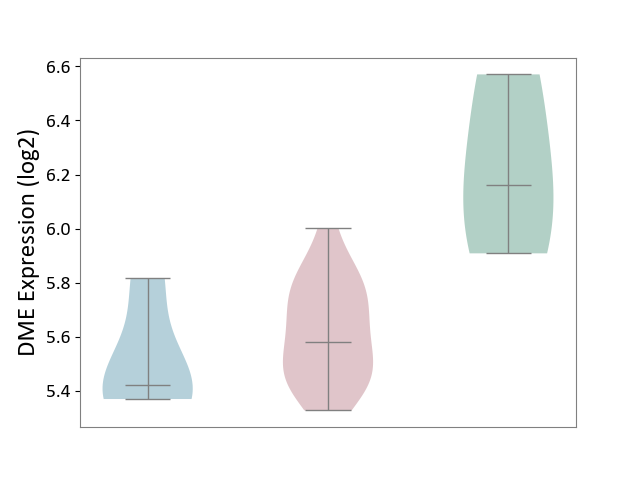
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 4A62 | Behcet disease | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Behcet's disease [ICD-11:4A62] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.42E-01; Fold-change: 6.96E-02; Z-score: 3.85E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
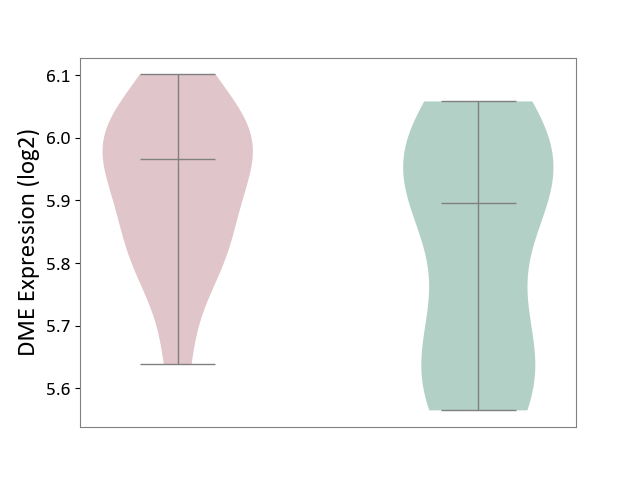
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 4B04 | Monocyte count disorder | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Autosomal dominant monocytopenia [ICD-11:4B04] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.44E-01; Fold-change: -2.77E-02; Z-score: -1.99E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
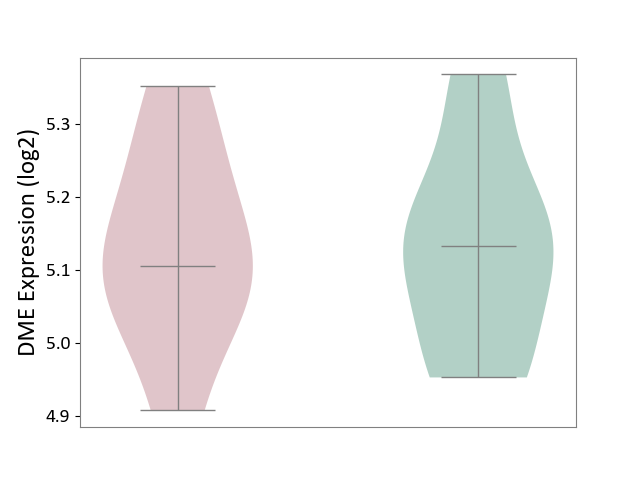
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 05 | Endocrine/nutritional/metabolic disease | Click to Show/Hide | |||
| ICD-11: 5A11 | Type 2 diabetes mellitus | Click to Show/Hide | |||
| The Studied Tissue | Omental adipose tissue | ||||
| The Specified Disease | Obesity related type 2 diabetes [ICD-11:5A11] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.74E-02; Fold-change: 2.10E-01; Z-score: 1.13E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
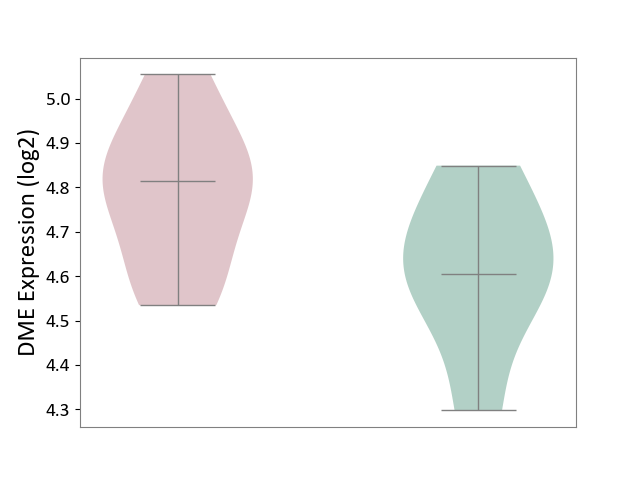
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Liver tissue | ||||
| The Specified Disease | Type 2 diabetes [ICD-11:5A11] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.04E-01; Fold-change: -2.88E-01; Z-score: -5.42E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
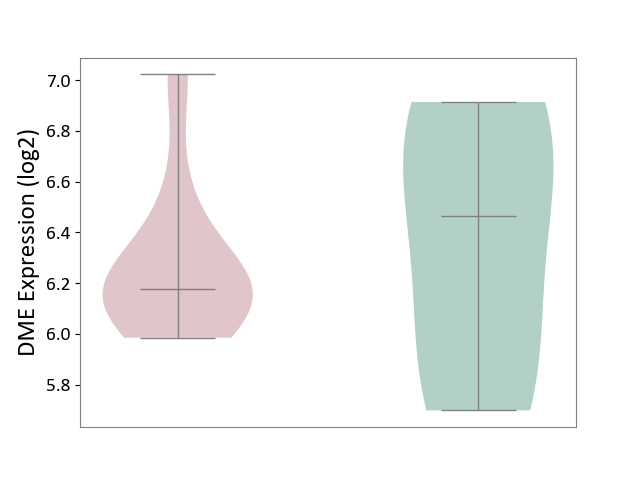
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 5A80 | Ovarian dysfunction | Click to Show/Hide | |||
| The Studied Tissue | Vastus lateralis muscle | ||||
| The Specified Disease | Polycystic ovary syndrome [ICD-11:5A80.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.22E-01; Fold-change: -3.99E-02; Z-score: -1.53E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
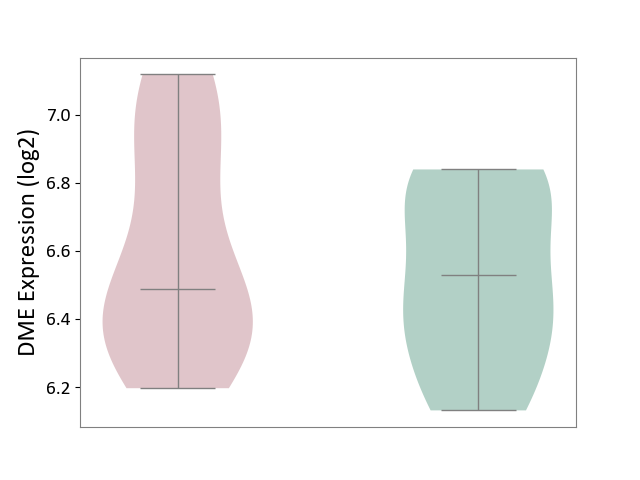
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 5C51 | Inborn carbohydrate metabolism disorder | Click to Show/Hide | |||
| The Studied Tissue | Biceps muscle | ||||
| The Specified Disease | Pompe disease [ICD-11:5C51.3] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.18E-01; Fold-change: -3.78E-02; Z-score: -2.13E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
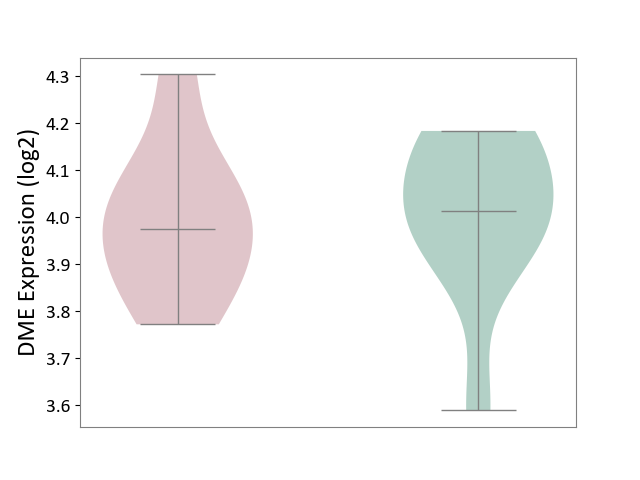
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 5C56 | Lysosomal disease | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Batten disease [ICD-11:5C56.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.03E-02; Fold-change: 7.98E-02; Z-score: 1.16E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
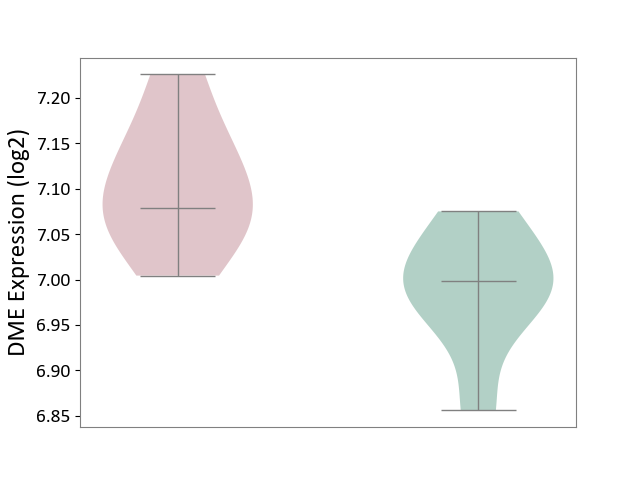
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 5C80 | Hyperlipoproteinaemia | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Familial hypercholesterolemia [ICD-11:5C80.00] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.10E-01; Fold-change: -5.43E-02; Z-score: -3.00E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
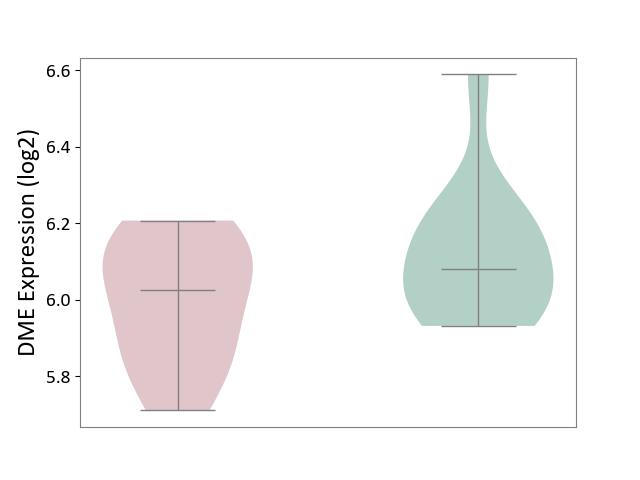
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Familial hypercholesterolemia [ICD-11:5C80.00] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.05E-04; Fold-change: -1.51E-01; Z-score: -8.87E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
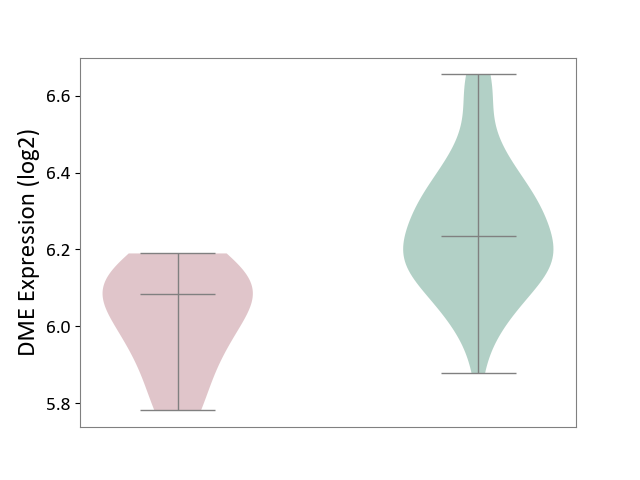
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 06 | Mental/behavioural/neurodevelopmental disorder | Click to Show/Hide | |||
| ICD-11: 6A02 | Autism spectrum disorder | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Autism [ICD-11:6A02] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.72E-01; Fold-change: 1.22E-01; Z-score: 4.54E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
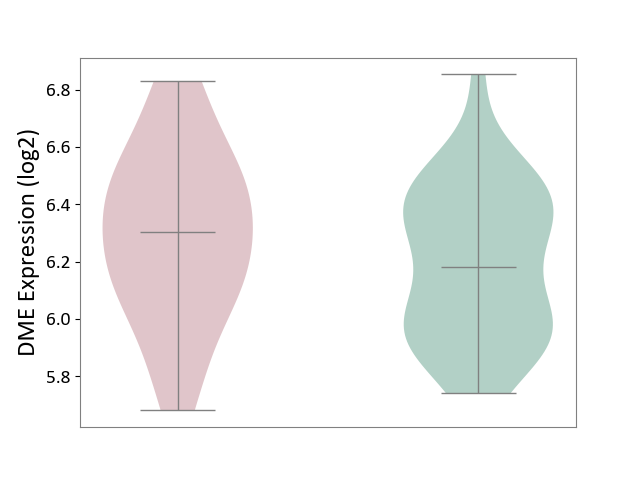
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 6A20 | Schizophrenia | Click to Show/Hide | |||
| The Studied Tissue | Prefrontal cortex | ||||
| The Specified Disease | Schizophrenia [ICD-11:6A20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.59E-01; Fold-change: 6.29E-02; Z-score: 3.14E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
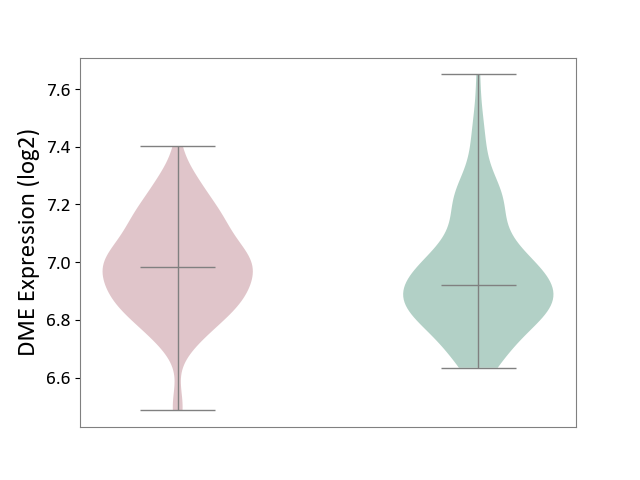
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Superior temporal cortex | ||||
| The Specified Disease | Schizophrenia [ICD-11:6A20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.84E-01; Fold-change: -8.38E-02; Z-score: -8.46E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
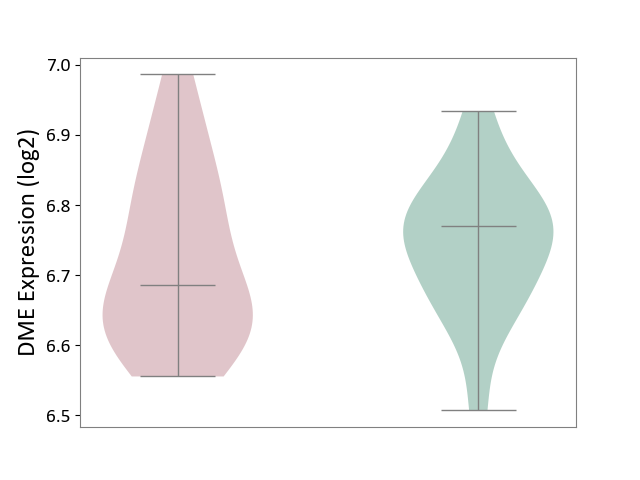
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 6A60 | Bipolar disorder | Click to Show/Hide | |||
| The Studied Tissue | Prefrontal cortex | ||||
| The Specified Disease | Bipolar disorder [ICD-11:6A60-6A6Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.18E-01; Fold-change: -4.95E-02; Z-score: -3.91E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
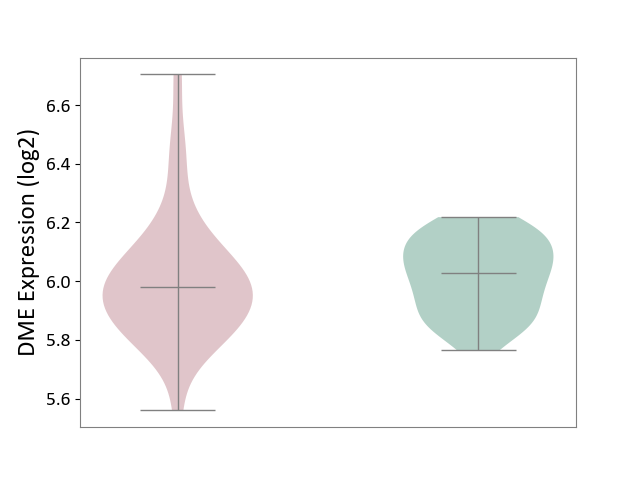
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 6A70 | Depressive disorder | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Major depressive disorder [ICD-11:6A70-6A7Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.65E-01; Fold-change: 4.83E-03; Z-score: 2.84E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
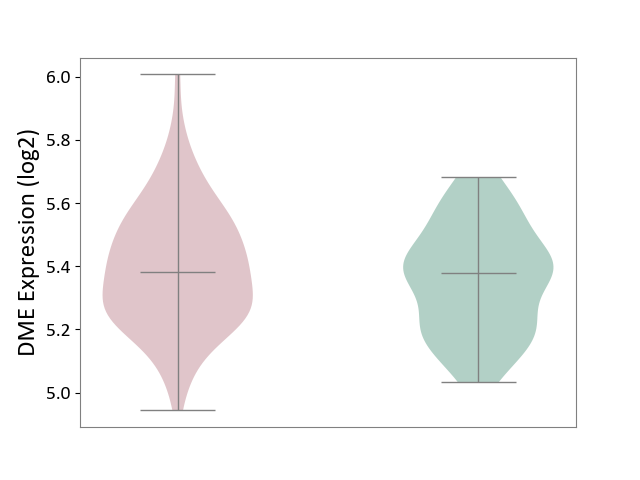
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Hippocampus | ||||
| The Specified Disease | Major depressive disorder [ICD-11:6A70-6A7Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.64E-01; Fold-change: 2.36E-02; Z-score: 1.75E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
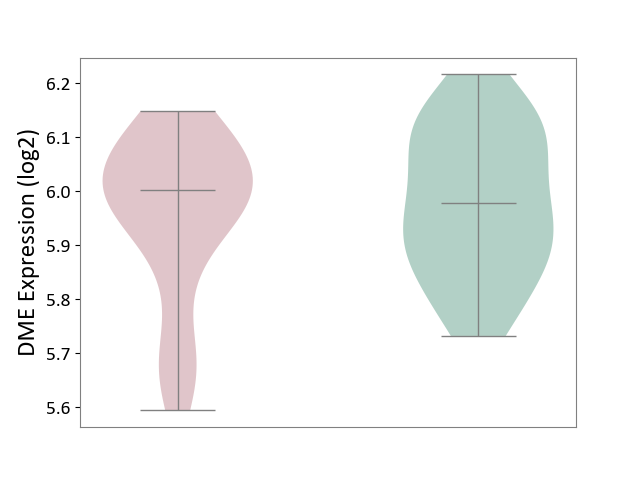
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 08 | Nervous system disease | Click to Show/Hide | |||
| ICD-11: 8A00 | Parkinsonism | Click to Show/Hide | |||
| The Studied Tissue | Substantia nigra tissue | ||||
| The Specified Disease | Parkinson's disease [ICD-11:8A00.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.70E-01; Fold-change: 2.98E-02; Z-score: 1.22E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
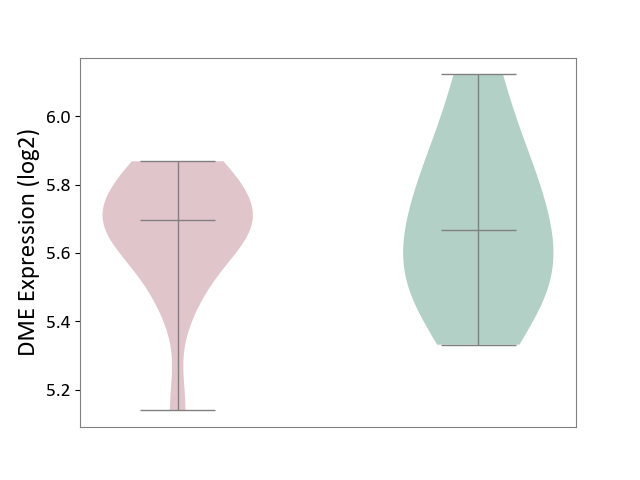
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8A01 | Choreiform disorder | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Huntington's disease [ICD-11:8A01.10] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.19E-01; Fold-change: 1.42E-01; Z-score: 7.09E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
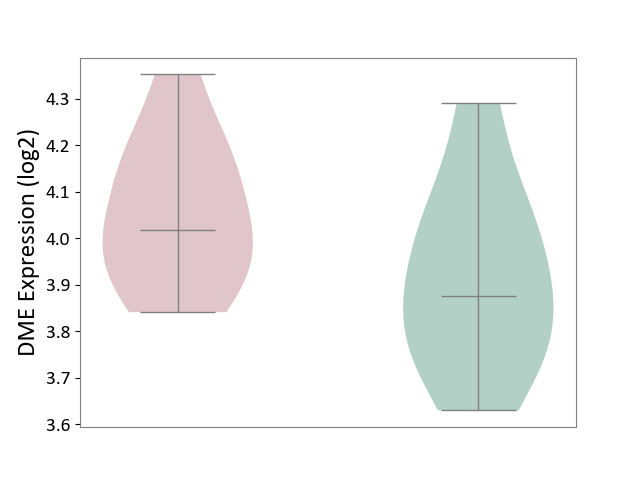
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8A20 | Alzheimer disease | Click to Show/Hide | |||
| The Studied Tissue | Entorhinal cortex | ||||
| The Specified Disease | Alzheimer's disease [ICD-11:8A20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.30E-01; Fold-change: -1.91E-02; Z-score: -1.41E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
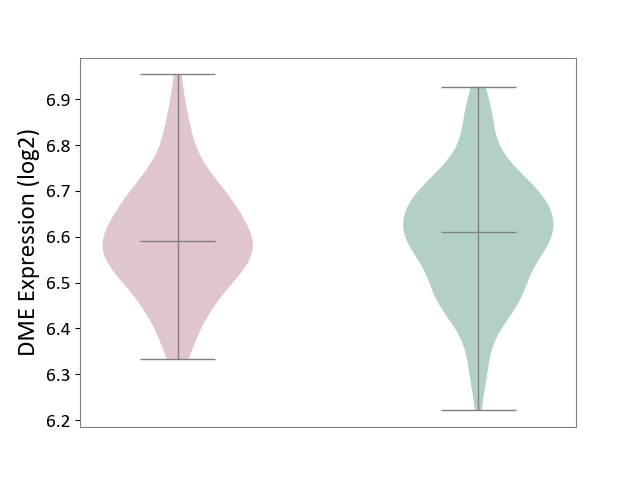
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8A2Y | Neurocognitive impairment | Click to Show/Hide | |||
| The Studied Tissue | White matter tissue | ||||
| The Specified Disease | HIV-associated neurocognitive impairment [ICD-11:8A2Y-8A2Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.56E-01; Fold-change: -6.08E-02; Z-score: -4.60E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
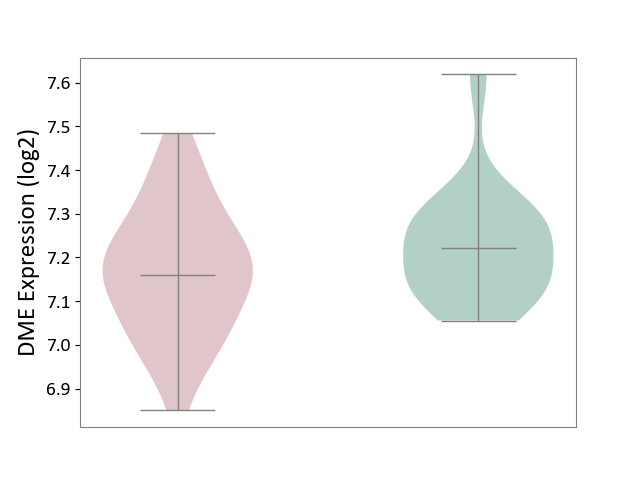
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8A40 | Multiple sclerosis | Click to Show/Hide | |||
| The Studied Tissue | Plasmacytoid dendritic cells | ||||
| The Specified Disease | Multiple sclerosis [ICD-11:8A40] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.95E-01; Fold-change: -7.47E-02; Z-score: -2.45E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
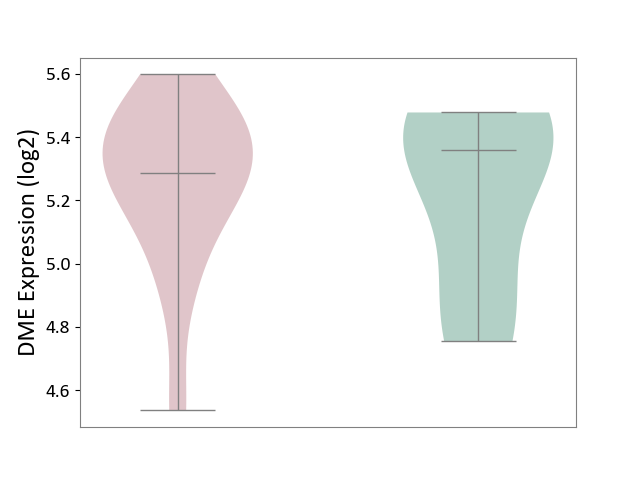
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Spinal cord | ||||
| The Specified Disease | Multiple sclerosis [ICD-11:8A40] | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.59E-02; Fold-change: -2.32E-02; Z-score: -1.99E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
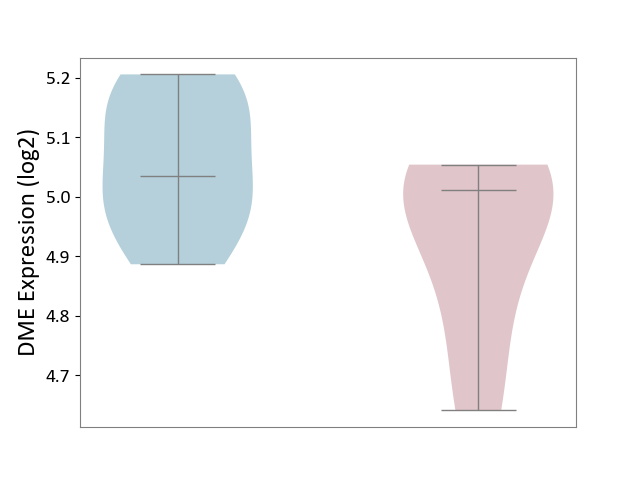
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8A60 | Epilepsy | Click to Show/Hide | |||
| The Studied Tissue | Peritumoral cortex tissue | ||||
| The Specified Disease | Epilepsy [ICD-11:8A60-8A6Z] | ||||
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 8.63E-01; Fold-change: 1.56E-01; Z-score: 6.73E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
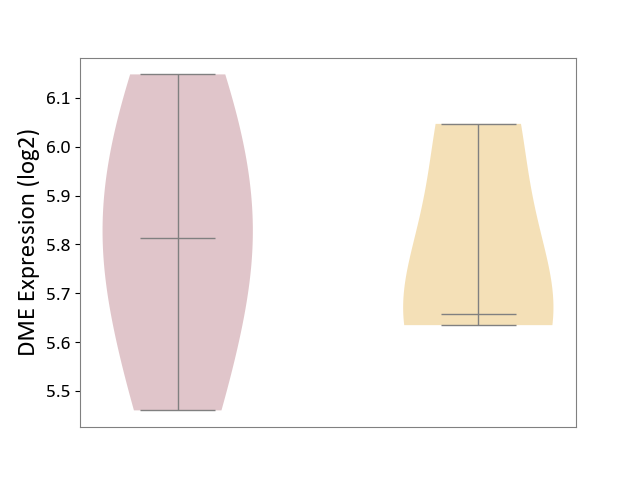
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Seizure [ICD-11:8A60-8A6Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.32E-01; Fold-change: 7.56E-02; Z-score: 4.21E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
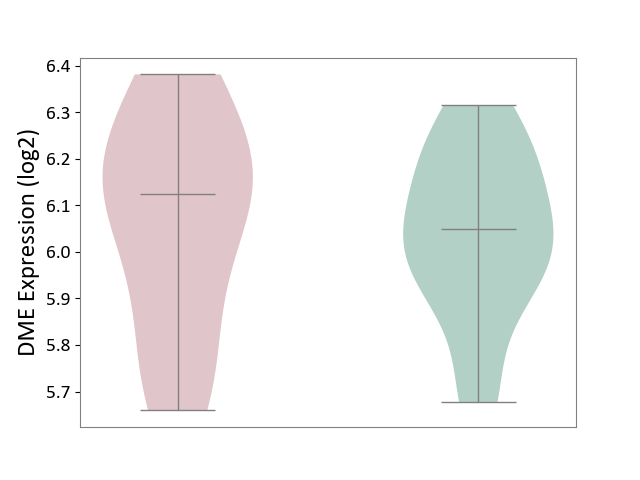
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8B01 | Subarachnoid haemorrhage | Click to Show/Hide | |||
| The Studied Tissue | Intracranial artery | ||||
| The Specified Disease | Intracranial aneurysm [ICD-11:8B01.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.54E-01; Fold-change: 8.93E-02; Z-score: 4.64E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
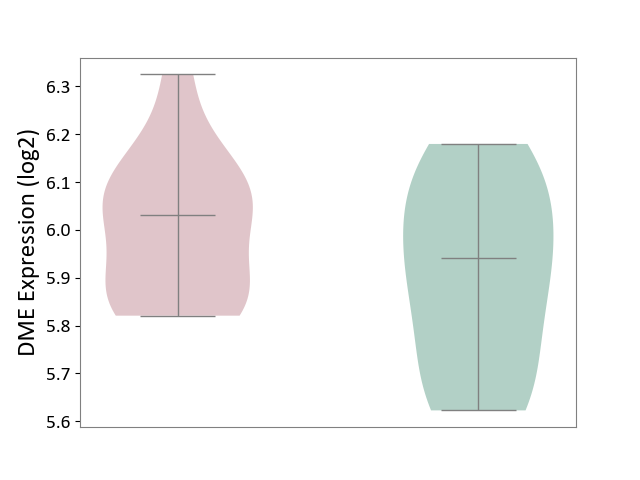
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8B11 | Cerebral ischaemic stroke | Click to Show/Hide | |||
| The Studied Tissue | Whole blood | ||||
| The Specified Disease | Cardioembolic stroke [ICD-11:8B11.20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.10E-01; Fold-change: -4.38E-02; Z-score: -3.62E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
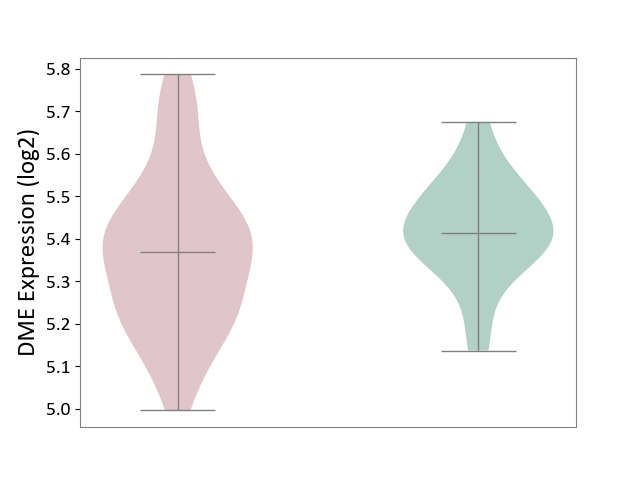
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Ischemic stroke [ICD-11:8B11] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-01; Fold-change: 6.24E-02; Z-score: 6.02E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
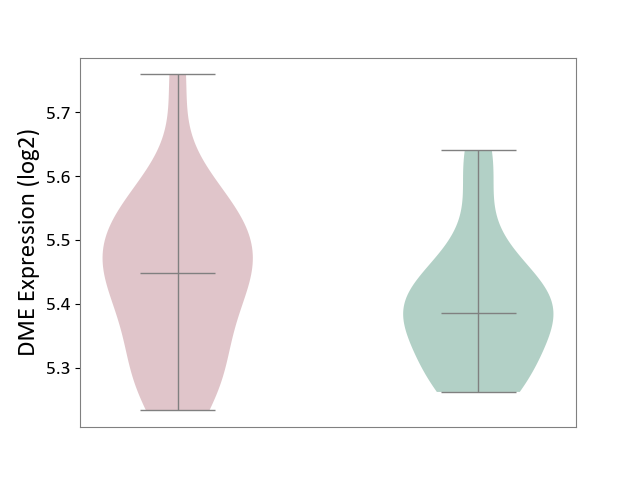
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8B60 | Motor neuron disease | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Lateral sclerosis [ICD-11:8B60.4] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.40E-01; Fold-change: 1.04E-01; Z-score: 7.11E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
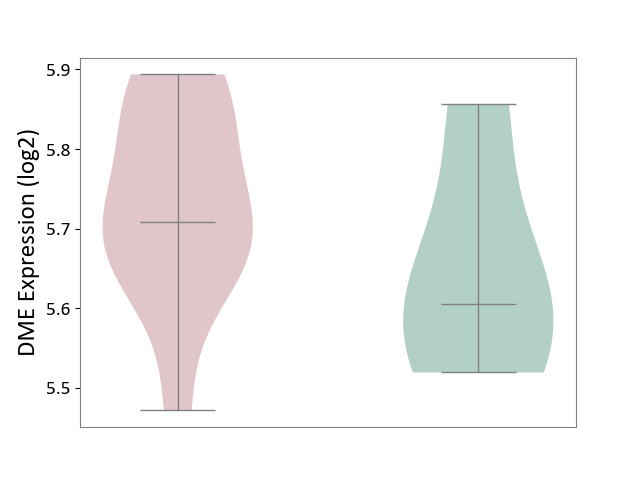
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Cervical spinal cord | ||||
| The Specified Disease | Lateral sclerosis [ICD-11:8B60.4] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.43E-01; Fold-change: 2.19E-03; Z-score: 1.72E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
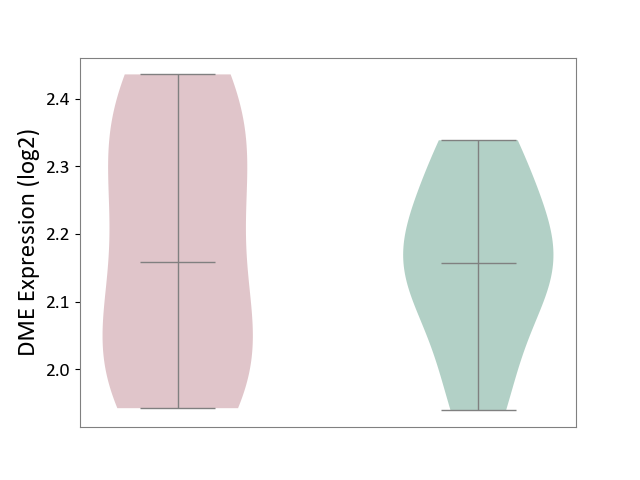
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8C70 | Muscular dystrophy | Click to Show/Hide | |||
| The Studied Tissue | Muscle tissue | ||||
| The Specified Disease | Myopathy [ICD-11:8C70.6] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.32E-03; Fold-change: -2.71E-01; Z-score: -1.58E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
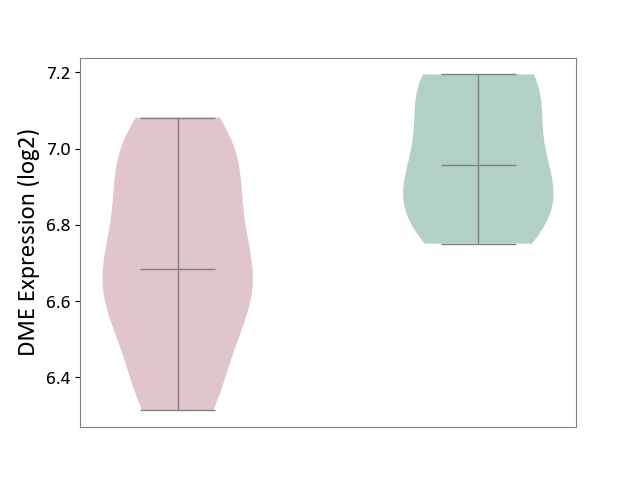
|
Click to View the Clearer Original Diagram | |||
| ICD-11: 8C75 | Distal myopathy | Click to Show/Hide | |||
| The Studied Tissue | Muscle tissue | ||||
| The Specified Disease | Tibial muscular dystrophy [ICD-11:8C75] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.51E-09; Fold-change: -6.44E-01; Z-score: -3.30E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
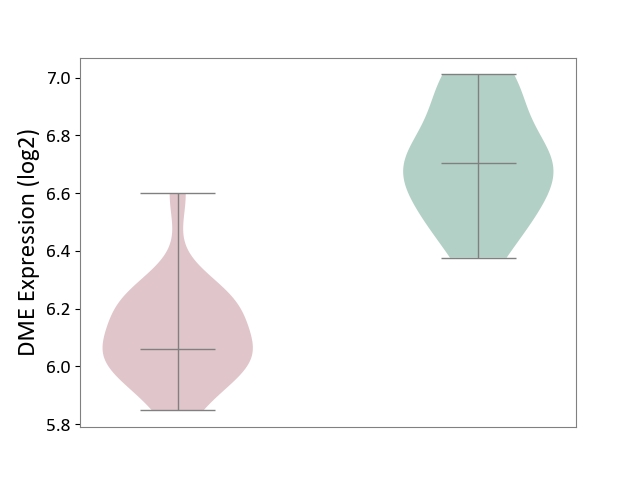
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 09 | Visual system disease | Click to Show/Hide | |||
| ICD-11: 9A96 | Anterior uveitis | Click to Show/Hide | |||
| The Studied Tissue | Peripheral monocyte | ||||
| The Specified Disease | Autoimmune uveitis [ICD-11:9A96] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.04E-03; Fold-change: -3.73E-01; Z-score: -1.91E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
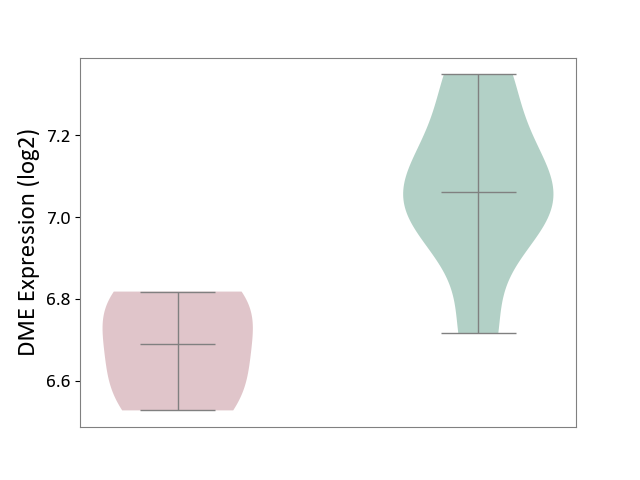
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 11 | Circulatory system disease | Click to Show/Hide | |||
| ICD-11: BA41 | Myocardial infarction | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Myocardial infarction [ICD-11:BA41-BA50] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.19E-02; Fold-change: 3.12E-01; Z-score: 8.06E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
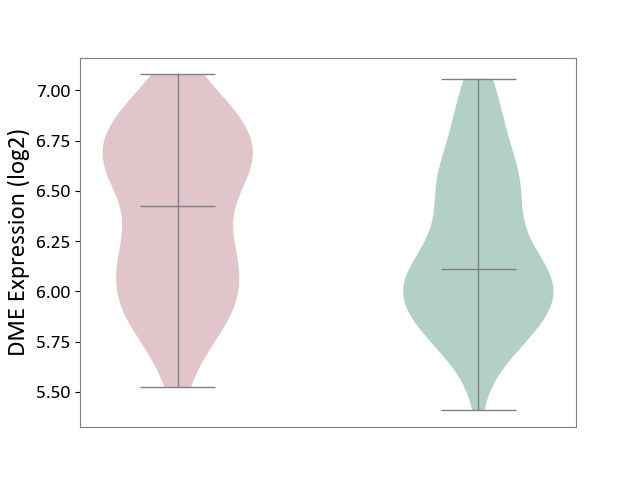
|
Click to View the Clearer Original Diagram | |||
| ICD-11: BA80 | Coronary artery disease | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Coronary artery disease [ICD-11:BA80-BA8Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.21E-01; Fold-change: -3.72E-02; Z-score: -1.96E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
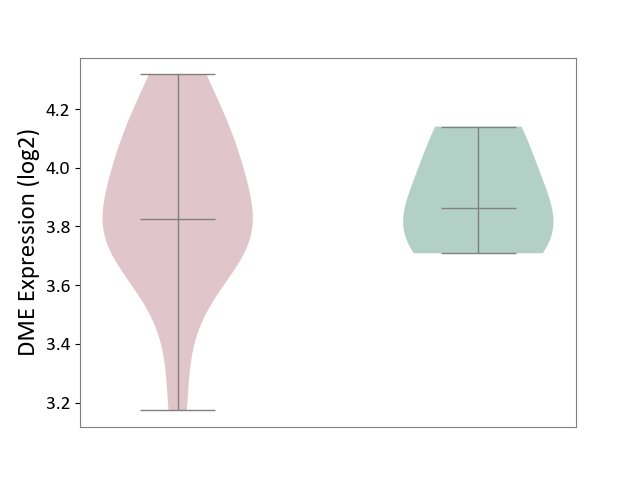
|
Click to View the Clearer Original Diagram | |||
| ICD-11: BB70 | Aortic valve stenosis | Click to Show/Hide | |||
| The Studied Tissue | Calcified aortic valve | ||||
| The Specified Disease | Aortic stenosis [ICD-11:BB70] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.67E-01; Fold-change: 1.35E-01; Z-score: 2.39E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
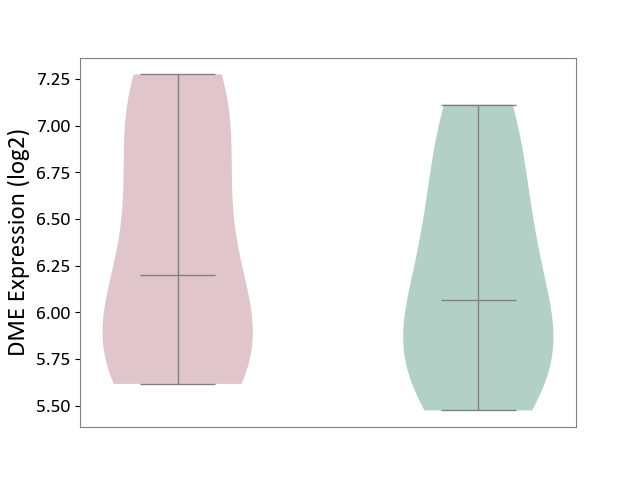
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 12 | Respiratory system disease | Click to Show/Hide | |||
| ICD-11: 7A40 | Central sleep apnoea | Click to Show/Hide | |||
| The Studied Tissue | Hyperplastic tonsil | ||||
| The Specified Disease | Apnea [ICD-11:7A40] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.31E-01; Fold-change: -1.20E-01; Z-score: -6.20E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
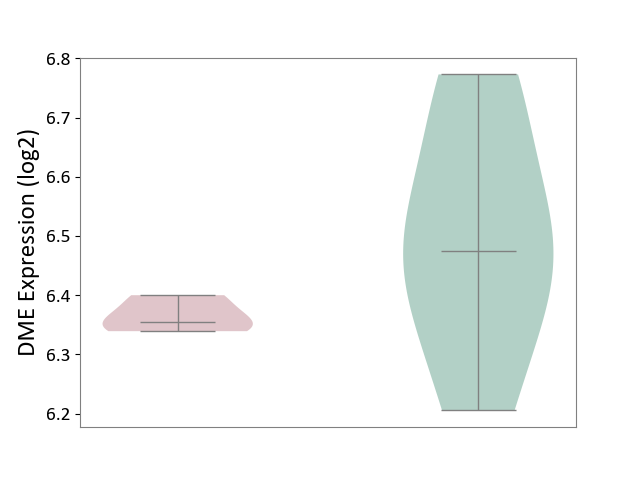
|
Click to View the Clearer Original Diagram | |||
| ICD-11: CA08 | Vasomotor or allergic rhinitis | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Olive pollen allergy [ICD-11:CA08.00] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.85E-01; Fold-change: -6.08E-02; Z-score: -2.34E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
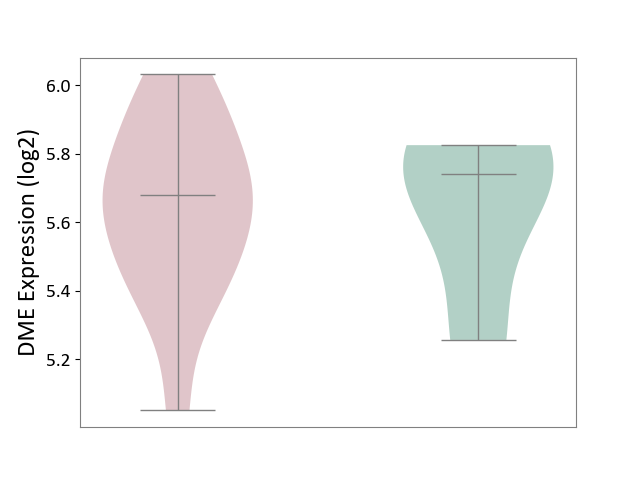
|
Click to View the Clearer Original Diagram | |||
| ICD-11: CA0A | Chronic rhinosinusitis | Click to Show/Hide | |||
| The Studied Tissue | Sinus mucosa tissue | ||||
| The Specified Disease | Chronic rhinosinusitis [ICD-11:CA0A] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.47E-01; Fold-change: -1.46E-01; Z-score: -1.13E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
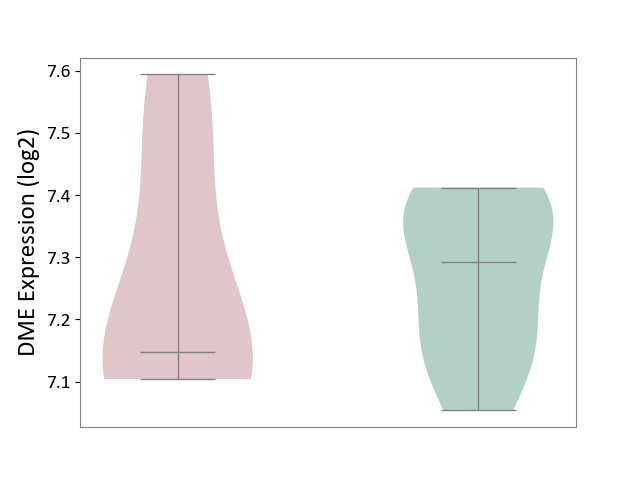
|
Click to View the Clearer Original Diagram | |||
| ICD-11: CA22 | Chronic obstructive pulmonary disease | Click to Show/Hide | |||
| The Studied Tissue | Lung tissue | ||||
| The Specified Disease | Chronic obstructive pulmonary disease [ICD-11:CA22] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.01E-02; Fold-change: 6.73E-02; Z-score: 3.36E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
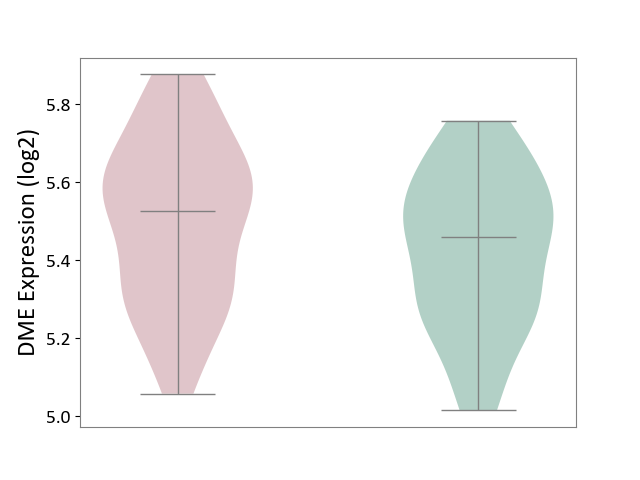
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Small airway epithelium | ||||
| The Specified Disease | Chronic obstructive pulmonary disease [ICD-11:CA22] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.91E-04; Fold-change: 1.41E-01; Z-score: 6.14E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
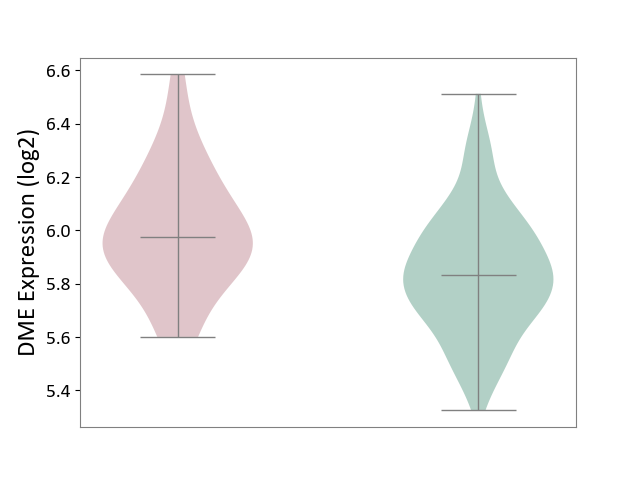
|
Click to View the Clearer Original Diagram | |||
| ICD-11: CA23 | Asthma | Click to Show/Hide | |||
| The Studied Tissue | Nasal and bronchial airway | ||||
| The Specified Disease | Asthma [ICD-11:CA23] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.21E-04; Fold-change: -2.33E-01; Z-score: -5.09E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
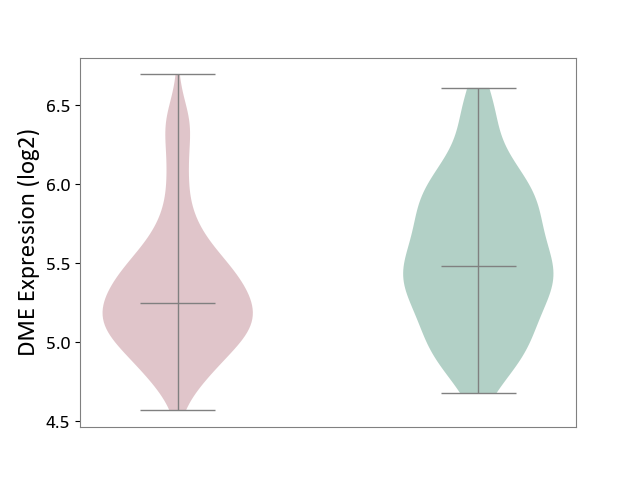
|
Click to View the Clearer Original Diagram | |||
| ICD-11: CB03 | Idiopathic interstitial pneumonitis | Click to Show/Hide | |||
| The Studied Tissue | Lung tissue | ||||
| The Specified Disease | Idiopathic pulmonary fibrosis [ICD-11:CB03.4] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.89E-01; Fold-change: -1.95E-01; Z-score: -9.15E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
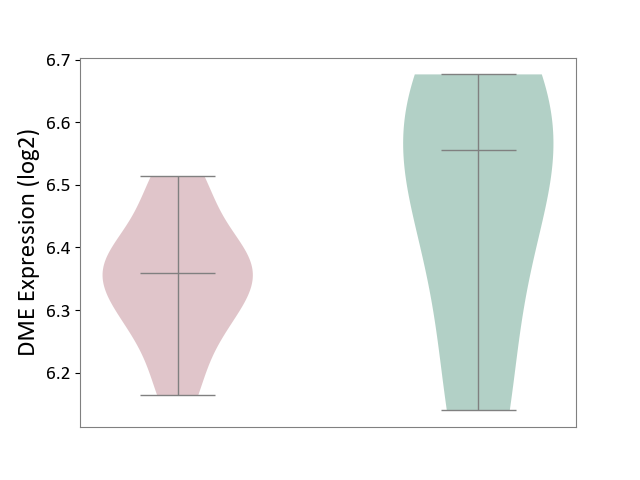
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 13 | Digestive system disease | Click to Show/Hide | |||
| ICD-11: DA0C | Periodontal disease | Click to Show/Hide | |||
| The Studied Tissue | Gingival tissue | ||||
| The Specified Disease | Periodontal disease [ICD-11:DA0C] | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.35E-02; Fold-change: 7.37E-02; Z-score: 3.65E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
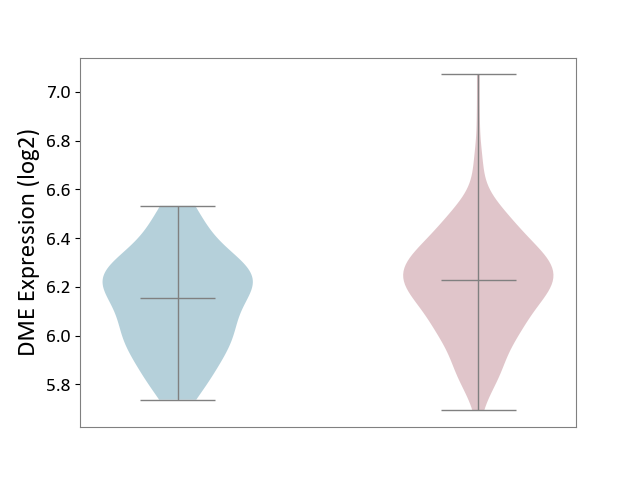
|
Click to View the Clearer Original Diagram | |||
| ICD-11: DA42 | Gastritis | Click to Show/Hide | |||
| The Studied Tissue | Gastric antrum tissue | ||||
| The Specified Disease | Eosinophilic gastritis [ICD-11:DA42.2] | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.90E-02; Fold-change: 1.81E-01; Z-score: 1.47E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
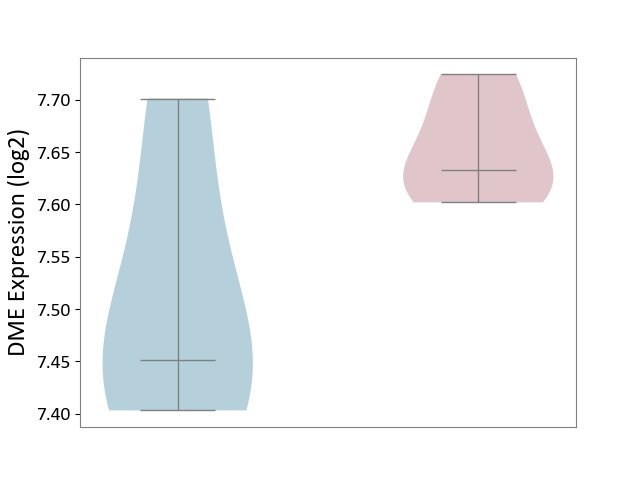
|
Click to View the Clearer Original Diagram | |||
| ICD-11: DB92 | Non-alcoholic fatty liver disease | Click to Show/Hide | |||
| The Studied Tissue | Liver tissue | ||||
| The Specified Disease | Non-alcoholic fatty liver disease [ICD-11:DB92] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.95E-01; Fold-change: -1.11E-01; Z-score: -5.80E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
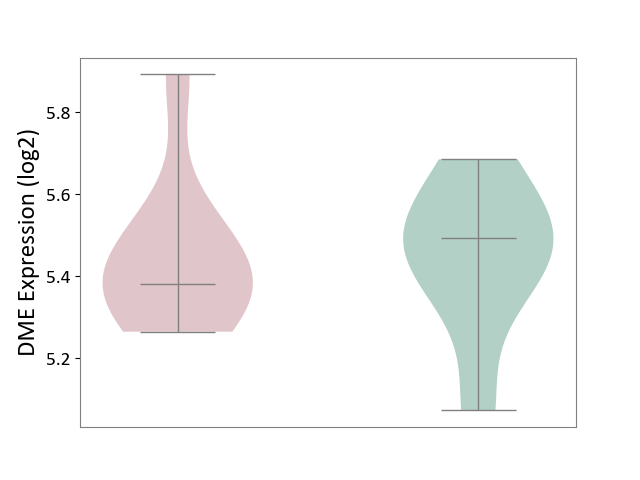
|
Click to View the Clearer Original Diagram | |||
| ICD-11: DB99 | Hepatic failure | Click to Show/Hide | |||
| The Studied Tissue | Liver tissue | ||||
| The Specified Disease | Liver failure [ICD-11:DB99.7-DB99.8] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.60E-02; Fold-change: -2.52E-01; Z-score: -1.10E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
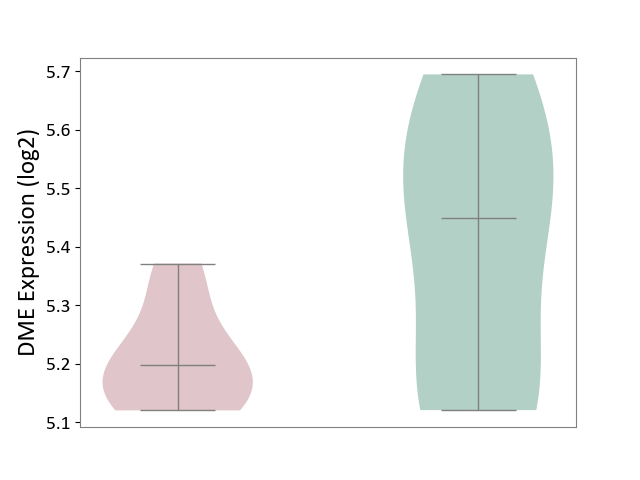
|
Click to View the Clearer Original Diagram | |||
| ICD-11: DD71 | Ulcerative colitis | Click to Show/Hide | |||
| The Studied Tissue | Colon mucosal tissue | ||||
| The Specified Disease | Ulcerative colitis [ICD-11:DD71] | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.70E-01; Fold-change: 1.35E-01; Z-score: 4.45E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in tissue other than the diseased tissue of patients
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
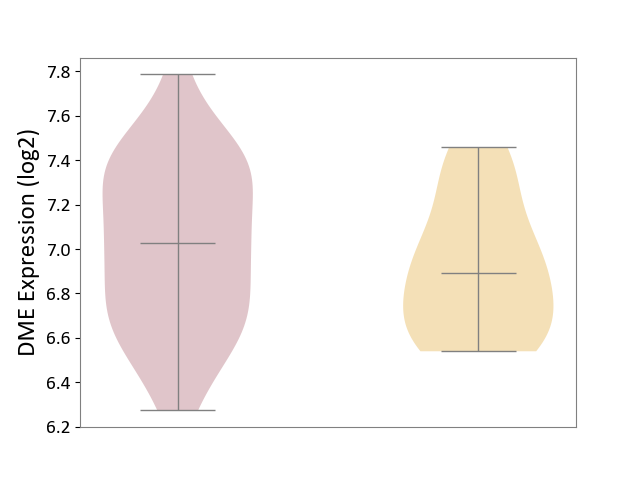
|
Click to View the Clearer Original Diagram | |||
| ICD-11: DD91 | Irritable bowel syndrome | Click to Show/Hide | |||
| The Studied Tissue | Rectal colon tissue | ||||
| The Specified Disease | Irritable bowel syndrome [ICD-11:DD91.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.20E-01; Fold-change: -1.63E-03; Z-score: -1.13E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
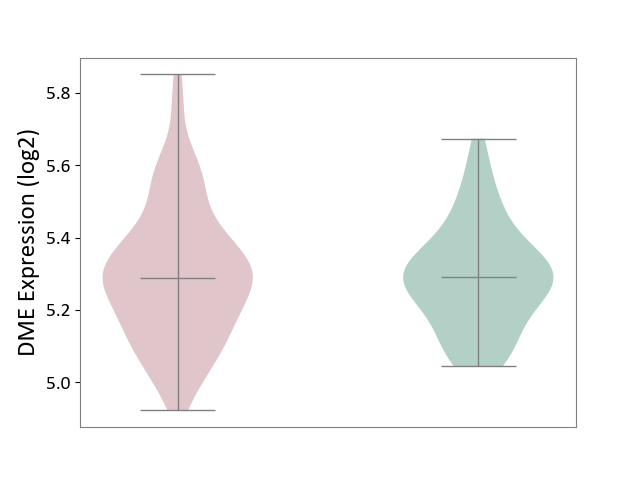
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 14 | Skin disease | Click to Show/Hide | |||
| ICD-11: EA80 | Atopic eczema | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Atopic dermatitis [ICD-11:EA80] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.35E-02; Fold-change: 2.21E-02; Z-score: 1.60E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
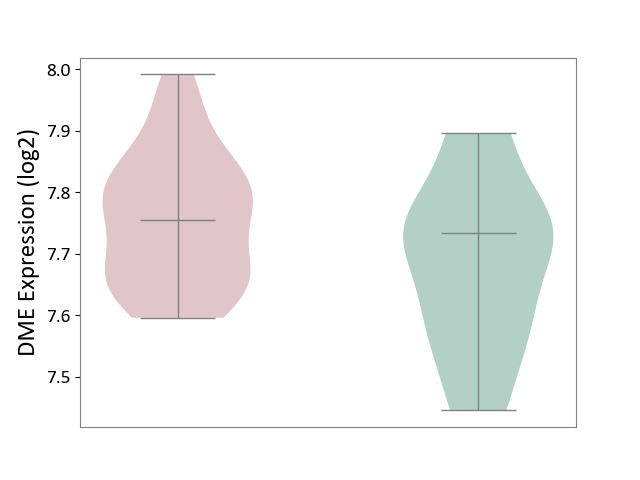
|
Click to View the Clearer Original Diagram | |||
| ICD-11: EA90 | Psoriasis | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Psoriasis [ICD-11:EA90] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.74E-01; Fold-change: -3.86E-02; Z-score: -1.04E-01 | ||||
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.54E-11; Fold-change: 3.09E-01; Z-score: 1.21E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue adjacent to the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
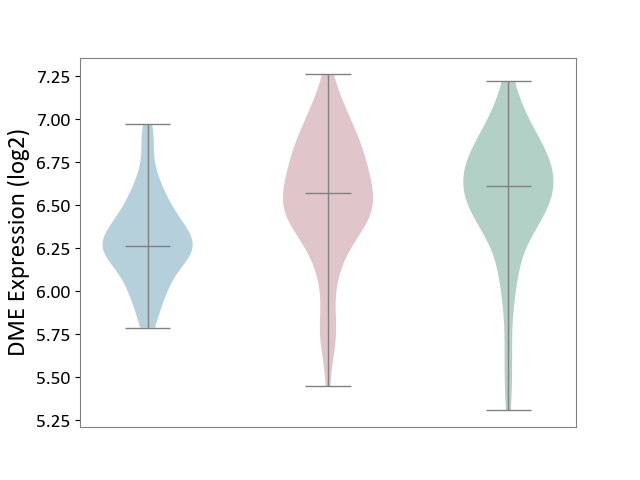
|
Click to View the Clearer Original Diagram | |||
| ICD-11: ED63 | Acquired hypomelanotic disorder | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Vitiligo [ICD-11:ED63.0] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.77E-01; Fold-change: -2.23E-02; Z-score: -1.96E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
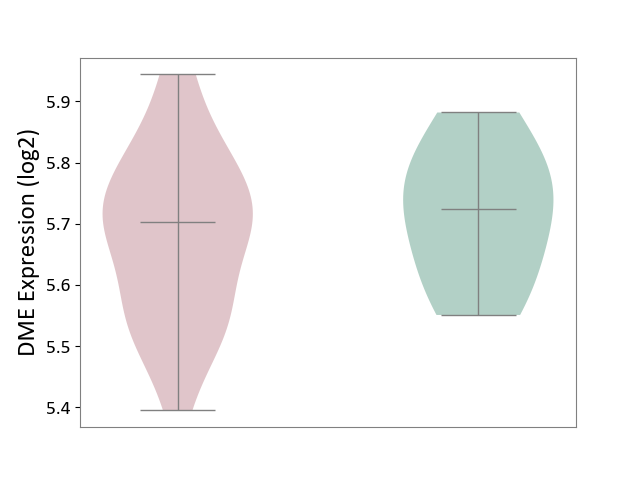
|
Click to View the Clearer Original Diagram | |||
| ICD-11: ED70 | Alopecia or hair loss | Click to Show/Hide | |||
| The Studied Tissue | Skin from scalp | ||||
| The Specified Disease | Alopecia [ICD-11:ED70] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.07E-01; Fold-change: -1.74E-02; Z-score: -7.66E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
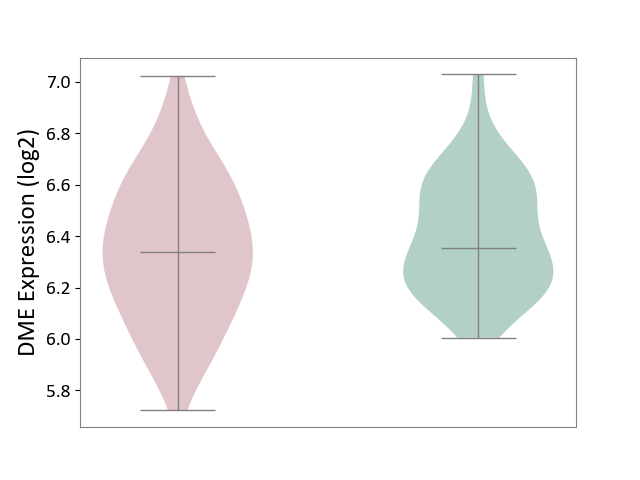
|
Click to View the Clearer Original Diagram | |||
| ICD-11: EK0Z | Contact dermatitis | Click to Show/Hide | |||
| The Studied Tissue | Skin | ||||
| The Specified Disease | Sensitive skin [ICD-11:EK0Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.28E-02; Fold-change: 7.52E-02; Z-score: 6.09E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
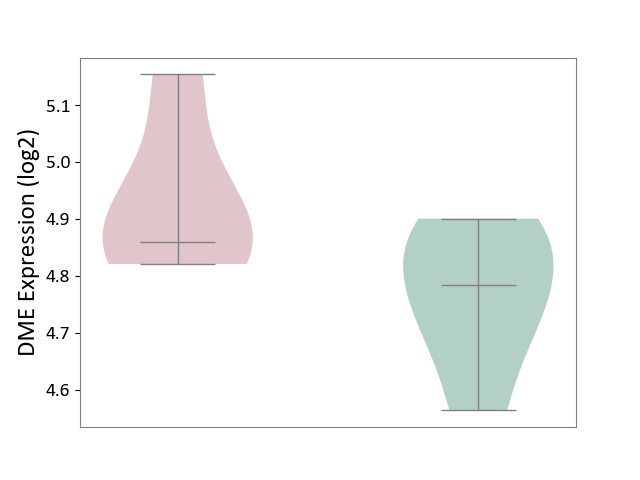
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 15 | Musculoskeletal system/connective tissue disease | Click to Show/Hide | |||
| ICD-11: FA00 | Osteoarthritis | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Arthropathy [ICD-11:FA00-FA5Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.39E-02; Fold-change: 3.35E-02; Z-score: 3.25E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
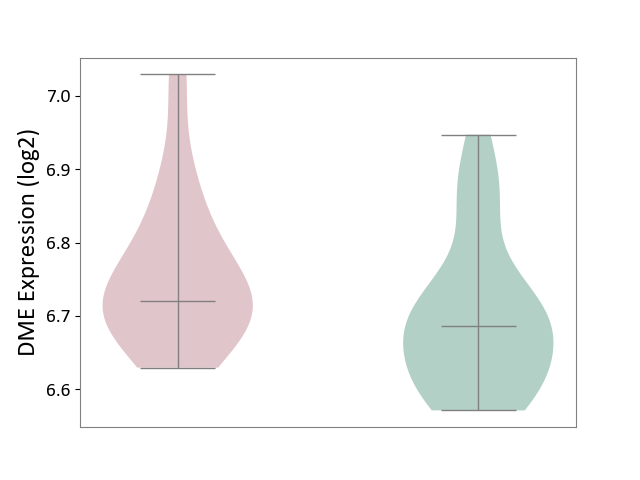
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Synovial tissue | ||||
| The Specified Disease | Osteoarthritis [ICD-11:FA00-FA0Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-01; Fold-change: -1.97E-01; Z-score: -7.83E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
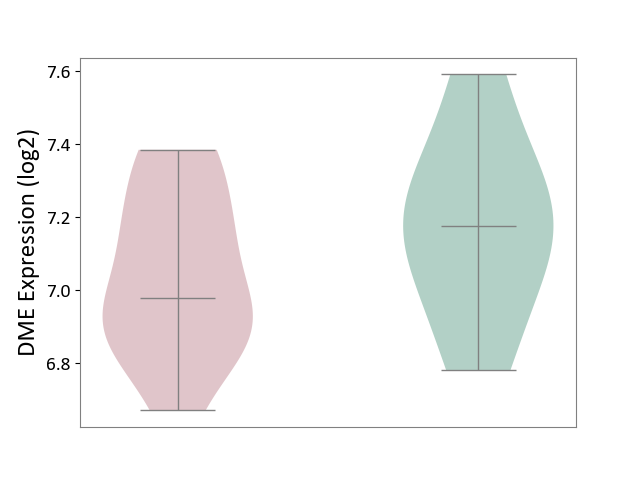
|
Click to View the Clearer Original Diagram | |||
| ICD-11: FA20 | Rheumatoid arthritis | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Childhood onset rheumatic disease [ICD-11:FA20.Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.46E-01; Fold-change: 3.70E-02; Z-score: 1.18E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
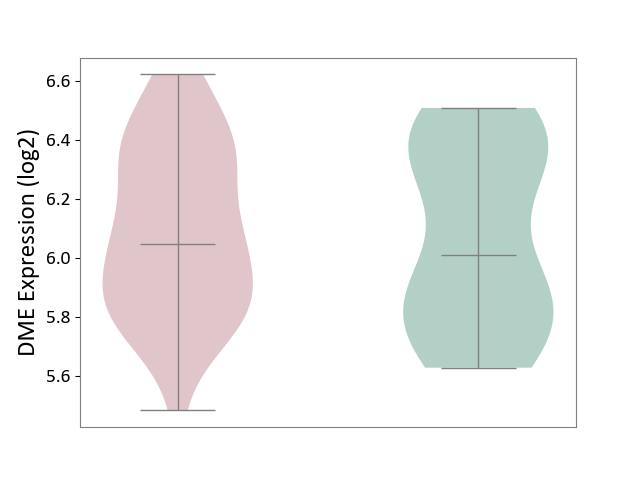
|
Click to View the Clearer Original Diagram | |||
| The Studied Tissue | Synovial tissue | ||||
| The Specified Disease | Rheumatoid arthritis [ICD-11:FA20] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.61E-03; Fold-change: -6.79E-01; Z-score: -2.18E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
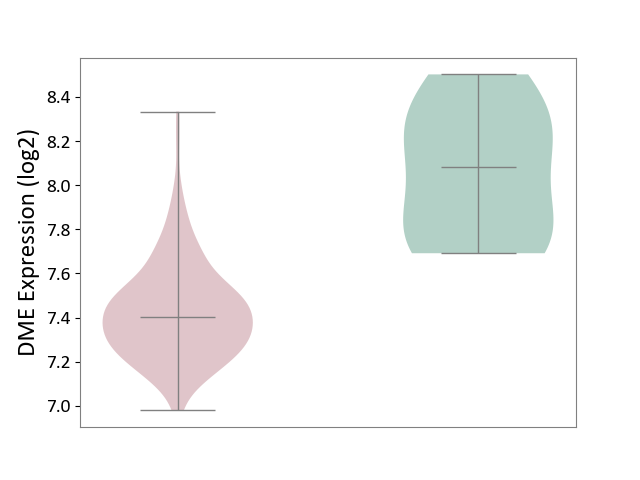
|
Click to View the Clearer Original Diagram | |||
| ICD-11: FA24 | Juvenile idiopathic arthritis | Click to Show/Hide | |||
| The Studied Tissue | Peripheral blood | ||||
| The Specified Disease | Juvenile idiopathic arthritis [ICD-11:FA24] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.87E-01; Fold-change: -3.81E-02; Z-score: -1.61E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
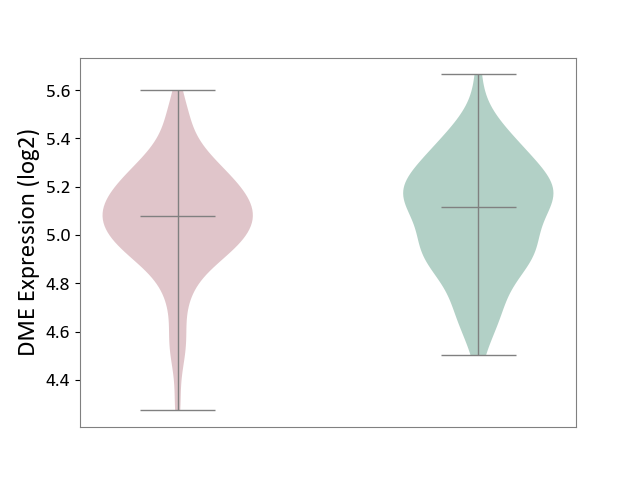
|
Click to View the Clearer Original Diagram | |||
| ICD-11: FA92 | Inflammatory spondyloarthritis | Click to Show/Hide | |||
| The Studied Tissue | Pheripheral blood | ||||
| The Specified Disease | Ankylosing spondylitis [ICD-11:FA92.0Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.53E-01; Fold-change: 1.21E-03; Z-score: 1.45E-02 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
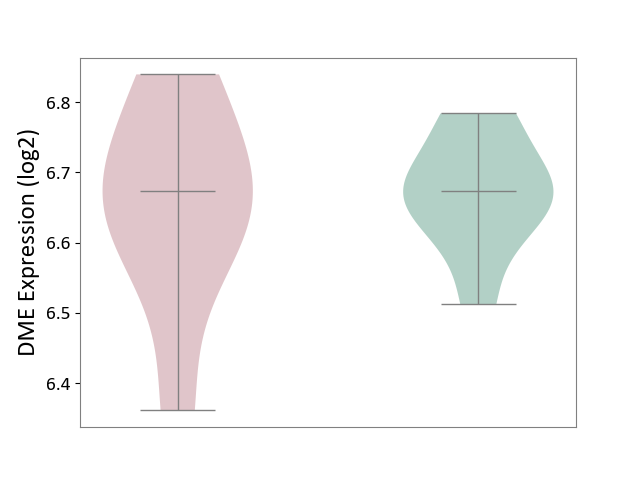
|
Click to View the Clearer Original Diagram | |||
| ICD-11: FB83 | Low bone mass disorder | Click to Show/Hide | |||
| The Studied Tissue | Bone marrow | ||||
| The Specified Disease | Osteoporosis [ICD-11:FB83.1] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.53E-04; Fold-change: 4.13E-01; Z-score: 3.30E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
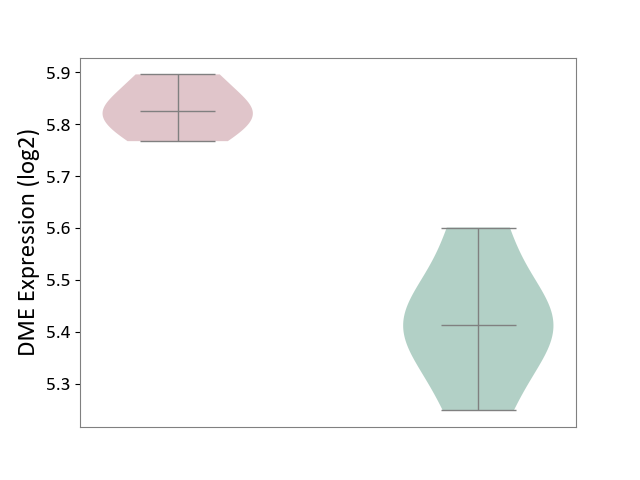
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 16 | Genitourinary system disease | Click to Show/Hide | |||
| ICD-11: GA10 | Endometriosis | Click to Show/Hide | |||
| The Studied Tissue | Endometrium tissue | ||||
| The Specified Disease | Endometriosis [ICD-11:GA10] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.63E-01; Fold-change: 8.24E-02; Z-score: 3.98E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
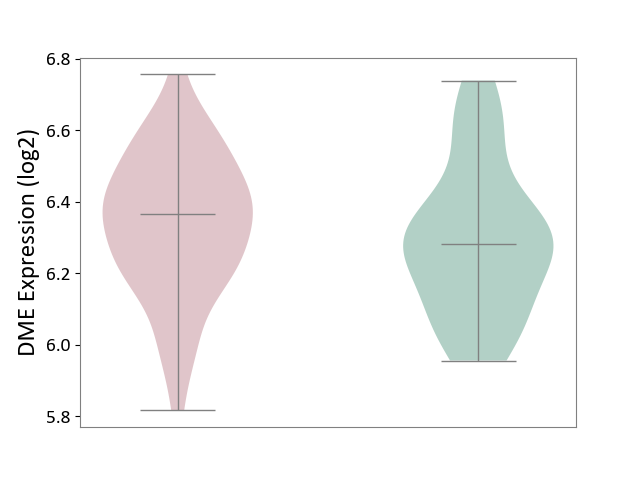
|
Click to View the Clearer Original Diagram | |||
| ICD-11: GC00 | Cystitis | Click to Show/Hide | |||
| The Studied Tissue | Bladder tissue | ||||
| The Specified Disease | Interstitial cystitis [ICD-11:GC00.3] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.22E-01; Fold-change: 6.96E-02; Z-score: 1.92E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
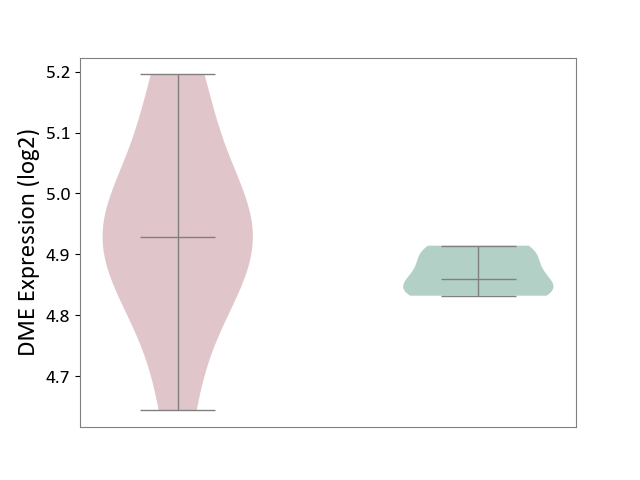
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 19 | Condition originating in perinatal period | Click to Show/Hide | |||
| ICD-11: KA21 | Short gestation disorder | Click to Show/Hide | |||
| The Studied Tissue | Myometrium | ||||
| The Specified Disease | Preterm birth [ICD-11:KA21.4Z] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.79E-01; Fold-change: -1.18E-01; Z-score: -8.68E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
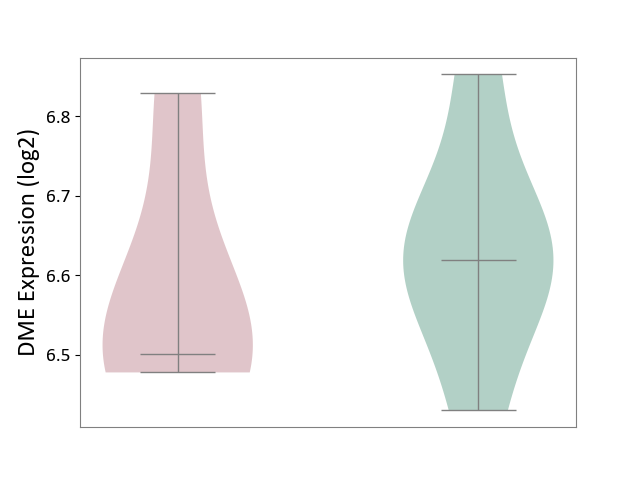
|
Click to View the Clearer Original Diagram | |||
| ICD Disease Classification 20 | Developmental anomaly | Click to Show/Hide | |||
| ICD-11: LD2C | Overgrowth syndrome | Click to Show/Hide | |||
| The Studied Tissue | Adipose tissue | ||||
| The Specified Disease | Simpson golabi behmel syndrome [ICD-11:LD2C] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.86E-01; Fold-change: -5.48E-02; Z-score: -4.55E-01 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
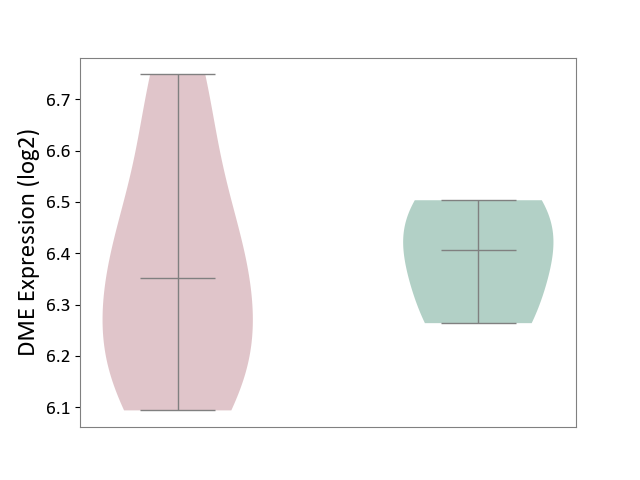
|
Click to View the Clearer Original Diagram | |||
| ICD-11: LD2D | Phakomatoses/hamartoneoplastic syndrome | Click to Show/Hide | |||
| The Studied Tissue | Perituberal tissue | ||||
| The Specified Disease | Tuberous sclerosis complex [ICD-11:LD2D.2] | ||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.79E-01; Fold-change: 8.11E-02; Z-score: 1.23E+00 | ||||
|
DME expression in the diseased tissue of patients
DME expression in the normal tissue of healthy individuals
|
|||||
| Violin Diagram of DME Disease-specific Protein Abundances |
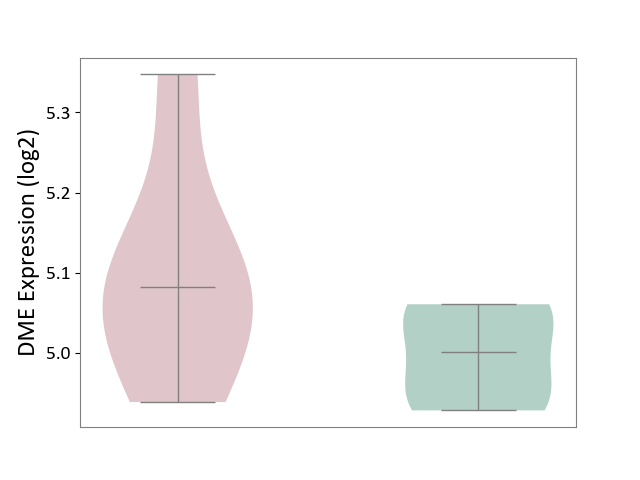
|
Click to View the Clearer Original Diagram | |||
| References | |||||
|---|---|---|---|---|---|
| 1 | Pharmacokinetics of bambuterol in subjects homozygous for the atypical gene for plasma cholinesterase | ||||
| 2 | Cobicistat versus ritonavir boosting and differences in the drug-drug interaction profiles with co-medications. J Antimicrob Chemother. 2016 Jul;71(7):1755-8. | ||||
| 3 | In vitro to in vivo extrapolation of the complex drug-drug interaction of bupropion and its metabolites with CYP2D6; simultaneous reversible inhibition and CYP2D6 downregulation. Biochem Pharmacol. 2017 Jan 1;123:85-96. | ||||
| 4 | CYP2D6 polymorphisms and tamoxifen metabolism: clinical relevance. Curr Oncol Rep. 2010 Jan;12(1):7-15. | ||||
| 5 | Metabolism and pharmacokinetics of novel selective vascular endothelial growth factor receptor-2 inhibitor apatinib in humans | ||||
| 6 | Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat Rev. 2009 Dec;35(8):692-706. | ||||
| 7 | Identification of Novel Pathways in Idelalisib Metabolism and Bioactivation | ||||
| 8 | DrugBank(Pharmacology-Metabolism)Estrone | ||||
| 9 | Metabolomics of Methylphenidate and Ethylphenidate: Implications in Pharmacological and Toxicological Effects | ||||
| 10 | Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch Toxicol. 1999 Mar;73(2):65-70. | ||||
| 11 | Neuronal cytochrome P450IID1 (debrisoquine/sparteine-type): potent inhibition of activity by (-)-cocaine and nucleotide sequence identity to human hepatic P450 gene CYP2D6. Mol Pharmacol. 1991 Jul;40(1):63-8. | ||||
| 12 | PubChem:Pimavanserin | ||||
| 13 | Substrates, inducers, inhibitors and structure-activity relationships of human Cytochrome P450 2C9 and implications in drug development. Curr Med Chem. 2009;16(27):3480-675. | ||||
| 14 | Inhibition of human cytochrome P450 isoforms by nonnucleoside reverse transcriptase inhibitors. J Clin Pharmacol. 2001 Jan;41(1):85-91. | ||||
| 15 | CYP2D6 and CYP3A4 involvement in the primary oxidative metabolism of hydrocodone by human liver microsomes. Br J Clin Pharmacol. 2004 Mar;57(3):287-97. | ||||
| 16 | Speculations on the substrate structure-activity relationship (SSAR) of cytochrome P450 enzymes. Biochem Pharmacol. 1992 Dec 1;44(11):2089-98. | ||||
| 17 | Characterization of In Vitro and In Vivo Metabolism of Antazoline Using Liquid Chromatography-Tandem Mass Spectrometry | ||||
| 18 | Clinical pharmacokinetics of galantamine. Clin Pharmacokinet. 2003;42(15):1383-92. | ||||
| 19 | Absorption, metabolism, and excretion of oral 14C radiolabeled ibrutinib: an open-label, phase I, single-dose study in healthy men. Drug Metab Dispos. 2015 Feb;43(2):289-97. | ||||
| 20 | Revisiting the metabolism of ketoconazole using accurate mass Drug Metab Lett. 2009 Aug;3(3):191-8. doi: 10.2174/187231209789352085. | ||||
| 21 | Metoclopramide is metabolized by CYP2D6 and is a reversible inhibitor, but not inactivator, of CYP2D6. Xenobiotica. 2014 Apr;44(4):309-319. | ||||
| 22 | Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur J Clin Pharmacol. 2003 Sep;59(5-6):429-42. | ||||
| 23 | Effect of genetic polymorphism on the metabolism of endogenous neuroactive substances, progesterone and p-tyramine, catalyzed by CYP2D6. Brain Res Mol Brain Res. 2004 Oct 22;129(1-2):117-23. | ||||
| 24 | Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr Med Chem. 2009;16(31):4066-218. | ||||
| 25 | Clinical pharmacokinetics of almotriptan, a serotonin 5-HT(1B/1D) receptor agonist for the treatment of migraine. Clin Pharmacokinet. 2005;44(3):237-46. | ||||
| 26 | Identification of the human liver enzymes involved in the metabolism of the antimigraine agent almotriptan. Drug Metab Dispos. 2003 Apr;31(4):404-11. | ||||
| 27 | Application of substrate depletion assay to evaluation of CYP isoforms responsible for stereoselective metabolism of carvedilol. Drug Metab Pharmacokinet. 2016 Dec;31(6):425-432. | ||||
| 28 | Multiple cytochrome P450 enzymes responsible for the oxidative metabolism of the substituted (S)-3-phenylpiperidine, (S,S)-3-[3-(methylsulfonyl)phenyl]-1-propylpiperidine hydrochloride, in human liver microsomes. Drug Metab Dispos. 2002 Dec;30(12):1372-7. | ||||
| 29 | Involvement of CYP1A2 and CYP3A4 in lidocaine N-deethylation and 3-hydroxylation in humans. Drug Metab Dispos. 2000 Aug;28(8):959-65. | ||||
| 30 | Exogenous cannabinoids as substrates, inhibitors, and inducers of human drug metabolizing enzymes: a systematic review. Drug Metab Rev. 2014 Feb;46(1):86-95. | ||||
| 31 | Metabolism of capsaicin by cytochrome P450 produces novel dehydrogenated metabolites and decreases cytotoxicity to lung and liver cells. Chem Res Toxicol. 2003 Mar;16(3):336-49. | ||||
| 32 | The biotransformation of clomipramine in vitro, identification of the cytochrome P450s responsible for the separate metabolic pathways. J Pharmacol Exp Ther. 1996 Jun;277(3):1659-64. | ||||
| 33 | Priapism induced by boceprevir-CYP3A4 inhibition and alpha-adrenergic blockade: case report. Clin Infect Dis. 2014 Jan;58(1):e35-8. | ||||
| 34 | CYP2C76-mediated species difference in drug metabolism: a comparison of pitavastatin metabolism between monkeys and humans | ||||
| 35 | Effect of health foods on cytochrome P450-mediated drug metabolism. J Pharm Health Care Sci. 2017 May 10;3:14. | ||||
| 36 | A phenotype-genotype approach to predicting CYP450 and P-glycoprotein drug interactions with the mixed inhibitor/inducer tipranavir/ritonavir. Clin Pharmacol Ther. 2010 Jun;87(6):735-42. | ||||
| 37 | Asenapine review, part I: chemistry, receptor affinity profile, pharmacokinetics and metabolism. Expert Opin Drug Metab Toxicol. 2014 Jun;10(6):893-903. | ||||
| 38 | Association of CYP2D6 and ADRB1 genes with hypotensive and antichronotropic action of betaxolol in patients with arterial hypertension. Fundam Clin Pharmacol. 2007 Aug;21(4):437-43. | ||||
| 39 | Oxidative metabolism of bupivacaine into pipecolylxylidine in humans is mainly catalyzed by CYP3A. Drug Metab Dispos. 2000 Apr;28(4):383-5. | ||||
| 40 | Pharmacogenomics knowledge for personalized medicine Clin Pharmacol Ther. 2012 Oct;92(4):414-7. doi: 10.1038/clpt.2012.96. | ||||
| 41 | Serum clomipramine and desmethylclomipramine levels in a CYP2C19 and CYP2D6 intermediate metabolizer. Pharmacogenomics. 2017 May;18(7):601-605. | ||||
| 42 | Erythromycin interaction with risperidone or clomipramine in an adolescent. J Child Adolesc Psychopharmacol. 1996 Summer;6(2):133-8. | ||||
| 43 | Identification of human liver cytochrome P450 isoforms involved in the in vitro metabolism of cyclobenzaprine. Drug Metab Dispos. 1996 Jul;24(7):786-91. | ||||
| 44 | Duloxetine: clinical pharmacokinetics and drug interactions. Clin Pharmacokinet. 2011 May;50(5):281-94. | ||||
| 45 | Comparison of verapamil, diltiazem, and labetalol on the bioavailability and metabolism of imipramine. J Clin Pharmacol. 1992 Feb;32(2):176-83. | ||||
| 46 | Role of cytochrome p450 isoenzymes 3A and 2D6 in the in vivo metabolism of mirabegron, a beta-3-adrenoceptor agonist. Clin Drug Investig. 2013 Jun;33(6):429-40. | ||||
| 47 | Metabolism and disposition of prescription opioids: a review. Forensic Sci Rev. 2015 Jul;27(2):115-45. | ||||
| 48 | Exposure to oral oxycodone is increased by concomitant inhibition of CYP2D6 and 3A4 pathways, but not by inhibition of CYP2D6 alone. Br J Clin Pharmacol. 2010 Jul;70(1):78-87. | ||||
| 49 | Ranolazine: a contemporary review. J Am Heart Assoc. 2016 Mar 15;5(3):e003196. | ||||
| 50 | In vitro characterization of the human biotransformation and CYP reaction phenotype of ET-743 (Yondelis, Trabectidin), a novel marine anti-cancer drug. Invest New Drugs. 2006 Jan;24(1):3-14. | ||||
| 51 | Genetic polymorphism of cytochrome P450 2D6 determines oestrogen receptor activity of the major infertility drug clomiphene via its active metabolites | ||||
| 52 | Effect of the CYP2D6*10 allele on the pharmacokinetics of clomiphene and its active metabolites | ||||
| 53 | Pharmacogenetics of schizophrenia. Am J Med Genet. 2000 Spring;97(1):98-106. | ||||
| 54 | Is therapeutic drug monitoring a case for optimizing clinical outcome and avoiding interactions of the selective serotonin reuptake inhibitors? Ther Drug Monit. 2000 Apr;22(2):143-54. | ||||
| 55 | Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002 Feb-May;34(1-2):83-448. | ||||
| 56 | The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014 Aug;21(8):879-85. | ||||
| 57 | Inhibition of desipramine hydroxylation (Cytochrome P450-2D6) in vitro by quinidine and by viral protease inhibitors: relation to drug interactions in vivo. J Pharm Sci. 1998 Oct;87(10):1184-9. | ||||
| 58 | Biocatalytic synthesis and structure elucidation of cyclized metabolites of the deacetylase inhibitor panobinostat (LBH589) Drug Metab Dispos. 2012 May;40(5):1041-50. doi: 10.1124/dmd.111.043620. | ||||
| 59 | Inhibition of cytochrome P450 2D6: structure-activity studies using a series of quinidine and quinine analogues. Chem Res Toxicol. 2003 Apr;16(4):450-9. | ||||
| 60 | Viral target and metabolism-based rationale for combined use of recently authorized small molecule COVID-19 medicines: Molnupiravir, nirmatrelvir, and remdesivir | ||||
| 61 | Identification of cytochrome P450 isozymes involved in metabolism of the alpha1-adrenoceptor blocker tamsulosin in human liver microsomes. Xenobiotica. 1998 Oct;28(10):909-22. | ||||
| 62 | Physiologically based pharmacokinetic (PBPK) modelling of tamsulosin related to CYP2D6*10 allele | ||||
| 63 | Brexpiprazole: first global approval. Drugs. 2015 Sep;75(14):1687-97. | ||||
| 64 | Determinants of interindividual variability and extent of CYP2D6 and CYP1A2 inhibition by paroxetine and fluvoxamine in vivo. J Clin Psychopharmacol. 1998 Jun;18(3):198-207. | ||||
| 65 | FDA Label of Deutetrabenazine. The 2020 official website of the U.S. Food and Drug Administration. | ||||
| 66 | Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet. 2009;48(11):689-723. | ||||
| 67 | Doxepin inhibits CYP2D6 activity in vivo. Pol J Pharmacol. 2004 Jul-Aug;56(4):491-4. | ||||
| 68 | Eliglustat: first global approval. Drugs. 2014 Oct;74(15):1829-36. | ||||
| 69 | FDA Approved Drug Products: Perforomist? inhalation solution | ||||
| 70 | Use of antidepressant drugs in schizophrenic patients with depression. Encephale. 2006 Mar-Apr;32(2 Pt 1):263-9. | ||||
| 71 | Disposition of lasofoxifene, a next-generation selective estrogen receptor modulator, in healthy male subjects | ||||
| 72 | DrugBank(Pharmacology-Metabolism):Lasofoxifene | ||||
| 73 | Identification of human cytochrome P450 and flavin-containing monooxygenase enzymes involved in the metabolism of lorcaserin, a novel selective human 5-hydroxytryptamine 2C agonist. Drug Metab Dispos. 2012 Apr;40(4):761-71. | ||||
| 74 | Influence of CYP2C9 and CYP2D6 polymorphisms on the pharmacokinetics of nateglinide in genotyped healthy volunteers. Clin Pharmacokinet. 2004;43(4):267-78. | ||||
| 75 | Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab Dispos. 1999 Dec;27(12):1488-95. | ||||
| 76 | Cytochrome P450 2D6 metabolism and 5-hydroxytryptamine type 3 receptor antagonists for postoperative nausea and vomiting. Med Sci Monit. 2005 Oct;11(10):RA322-8. | ||||
| 77 | Population pharmacokinetics of pomalidomide. J Clin Pharmacol. 2015 May;55(5):563-72. | ||||
| 78 | Effect of selective serotonin reuptake inhibitors on the oxidative metabolism of propafenone: in vitro studies using human liver microsomes. J Clin Psychopharmacol. 2000 Aug;20(4):428-34. | ||||
| 79 | Structure-activity relationship for human cytochrome P450 substrates and inhibitors. Drug Metab Rev. 2002 Feb-May;34(1-2):69-82. | ||||
| 80 | Evaluating the role of drug metabolism and reactive intermediates in trazodone-induced cytotoxicity toward freshly-isolated rat hepatocytes. Drug Res (Stuttg). 2016 Nov;66(11):592-596. | ||||
| 81 | The combination of umeclidinium bromide and vilanterol in the management of chronic obstructive pulmonary disease: current evidence and future prospects. Ther Adv Respir Dis. 2013 Dec;7(6):311-9. | ||||
| 82 | Bioactivation of the tricyclic antidepressant amitriptyline and its metabolite nortriptyline to arene oxide intermediates in human liver microsomes and recombinant P450s. Chem Biol Interact. 2008 May 9;173(1):59-67. | ||||
| 83 | Amprenavir: a new human immunodeficiency virus type 1 protease inhibitor. Clin Ther. 2000 May;22(5):549-72. | ||||
| 84 | The contribution of the enzymes CYP2D6 and CYP2C19 in the demethylation of artemether in healthy subjects. Eur J Drug Metab Pharmacokinet. 1998 Jul-Sep;23(3):429-36. | ||||
| 85 | Atomoxetine: a review of its pharmacokinetics and pharmacogenomics relative to drug disposition. J Child Adolesc Psychopharmacol. 2016 May;26(4):314-26. | ||||
| 86 | Transport, metabolism, and in vivo population pharmacokinetics of the chloro benztropine analogs, a class of compounds extensively evaluated in animal models of drug abuse. J Pharmacol Exp Ther. 2007 Jan;320(1):344-53. | ||||
| 87 | DrugBank(Pharmacology-Metabolism):Betrixaban | ||||
| 88 | Metabolite Profiling and Reaction Phenotyping for the in Vitro Assessment of the Bioactivation of Bromfenac ? Chem Res Toxicol. 2020 Jan 21;33(1):249-257. doi: 10.1021/acs.chemrestox.9b00268. | ||||
| 89 | Cytochrome P450 3A-mediated metabolism of buspirone in human liver microsomes. Drug Metab Dispos. 2005 Apr;33(4):500-7. | ||||
| 90 | Pharmacokinetics and metabolism of the novel muscarinic receptor agonist SNI-2011 in rats and dogs. Arzneimittelforschung. 2003;53(1):26-33. | ||||
| 91 | Clinical pharmacokinetic and pharmacodynamic profile of cinacalcet hydrochloride. Clin Pharmacokinet. 2009;48(5):303-11. | ||||
| 92 | The clinical pharmacokinetics of darifenacin. Clin Pharmacokinet. 2006;45(4):325-50. | ||||
| 93 | Effect of the CYP2D6*10 genotype on venlafaxine pharmacokinetics in healthy adult volunteers. Br J Clin Pharmacol. 1999 Apr;47(4):450-3. | ||||
| 94 | Drug interactions with calcium channel blockers: possible involvement of metabolite-intermediate complexation with CYP3A. Drug Metab Dispos. 2000 Feb;28(2):125-30. | ||||
| 95 | Donepezil plasma concentrations, CYP2D6 and CYP3A4 phenotypes, and cognitive outcome in Alzheimer's disease. Eur J Clin Pharmacol. 2016 Jun;72(6):711-7. | ||||
| 96 | Inhibitory effects of anticancer drugs on dextromethorphan-O-demethylase activity in human liver microsomes. Cancer Chemother Pharmacol. 1993;32(6):491-5. | ||||
| 97 | Effect of dronedarone on the pharmacokinetics of carvedilol following oral administration to rats. Eur J Pharm Sci. 2018 Jan 1;111:13-19. | ||||
| 98 | Pharmacokinetics, metabolism, and disposition of deferasirox in beta-thalassemic patients with transfusion-dependent iron overload who are at pharmacokinetic steady state Drug Metab Dispos. 2010 May;38(5):808-16. doi: 10.1124/dmd.109.030833. | ||||
| 99 | (R)-, (S)-, and racemic fluoxetine N-demethylation by human cytochrome P450 enzymes. Drug Metab Dispos. 2000 Oct;28(10):1187-91. | ||||
| 100 | Clinical pharmacokinetics of fluvastatin. Clin Pharmacokinet. 2001;40(4):263-81. | ||||
| 101 | QTc prolongation associated with combination therapy of levofloxacin, imipramine, and fluoxetine. Ann Pharmacother. 2005 Mar;39(3):543-6. | ||||
| 102 | Contribution of UDP-glucuronosyltransferases 1A9 and 2B7 to the glucuronidation of indomethacin in the human liver. Eur J Clin Pharmacol. 2007 Mar;63(3):289-96. | ||||
| 103 | Effects of rifampin on the pharmacokinetics of a single dose of istradefylline in healthy subjects. J Clin Pharmacol. 2018 Feb;58(2):193-201. | ||||
| 104 | Characterization of Stereoselective Metabolism, Inhibitory Effect on Uric Acid Uptake Transporters, and Pharmacokinetics of Lesinurad Atropisomers Drug Metab Dispos. 2019 Feb;47(2):104-113. doi: 10.1124/dmd.118.080549. | ||||
| 105 | LABEL:PREVYMIS- letermovir tablet, film coated PREVYMIS- letermovir injection, solution | ||||
| 106 | Influence of the CYP2D6 isoenzyme in patients treated with venlafaxine for major depressive disorder: clinical and economic consequences. PLoS One. 2014 Nov 4;9(11):e90453. | ||||
| 107 | Preclinical pharmacokinetics, pharmacology and toxicology of lisdexamfetamine: a novel d-amphetamine pro-drug. Neuropharmacology. 2014 Dec;87:41-50. | ||||
| 108 | The impact of CYP2D6 mediated drug-drug interaction: a systematic review on a combination of metoprolol and paroxetine/fluoxetine. Br J Clin Pharmacol. 2018 Dec;84(12):2704-2715. | ||||
| 109 | Activation of G-proteins by morphine and codeine congeners: insights to the relevance of O- and N-demethylated metabolites at mu- and delta-opioid receptors. J Pharmacol Exp Ther. 2004 Feb;308(2):547-54. | ||||
| 110 | Nefazodone, meta-chlorophenylpiperazine, and their metabolites in vitro: cytochromes mediating transformation, and P450-3A4 inhibitory actions. Psychopharmacology (Berl). 1999 Jul;145(1):113-22. | ||||
| 111 | [Olanzapine: pharmacology, pharmacokinetics and therapeutic drug monitoring]. Fortschr Neurol Psychiatr. 2001 Nov;69(11):510-7. | ||||
| 112 | Binding of quinidine radically increases the stability and decreases the flexibility of the cytochrome P450 2D6 active site. J Inorg Biochem. 2012 May;110:46-50. | ||||
| 113 | A 28-day, randomized, double-blind, placebo-controlled, parallel group study of nebulized revefenacin in patients with chronic obstructive pulmonary disease. Respir Res. 2017 Nov 2;18(1):182. | ||||
| 114 | Evaluation of absorption, distribution, metabolism, and excretion of [(14)C]-rucaparib, a poly(ADP-ribose) polymerase inhibitor, in patients with advanced solid tumors Invest New Drugs. 2020 Jun;38(3):765-775. doi: 10.1007/s10637-019-00815-2. | ||||
| 115 | Genetic polymorphisms in cytochrome P450 enzymes: effect on efficacy and tolerability of HMG-CoA reductase inhibitors. Am J Cardiovasc Drugs. 2004;4(4):247-55. | ||||
| 116 | Investigations into the drug-drug interaction potential of tapentadol in human liver microsomes and fresh human hepatocytes. Drug Metab Lett. 2008 Jan;2(1):67-75. | ||||
| 117 | In vitro metabolism of tegaserod in human liver and intestine: assessment of drug interactions. Drug Metab Dispos. 2001 Oct;29(10):1269-76. | ||||
| 118 | Effect of zolpidem on human cytochrome P450 activity, and on transport mediated by P-glycoprotein. Biopharm Drug Dispos. 2002 Dec;23(9):361-7. | ||||
| 119 | Potentially significant drug interactions of class III antiarrhythmic drugs. Drug Saf. 2003;26(6):421-38. | ||||
| 120 | Addition of amoxapine improves positive and negative symptoms in a patient with schizophrenia Ther Adv Psychopharmacol. 2013 Dec;3(6):340-2. doi: 10.1177/2045125313499363. | ||||
| 121 | Inhibitory effects of azelastine and its metabolites on drug oxidation catalyzed by human cytochrome P-450 enzymes. Drug Metab Dispos. 1999 Jul;27(7):792-7. | ||||
| 122 | Effect of nonspecific binding to microsomes and metabolic elimination of buprenorphine on the inhibition of cytochrome P4502D6. Biol Pharm Bull. 2005 Feb;28(2):212-6. | ||||
| 123 | Pharmacokinetic-pharmacodynamic relationship of the selective serotonin reuptake inhibitors. Clin Pharmacokinet. 1996 Dec;31(6):444-69. | ||||
| 124 | CYP2D6 mediates 4-hydroxylation of clonidine in vitro: implication for pregnancy-induced changes in clonidine clearance. Drug Metab Dispos. 2010 Sep;38(9):1393-6. | ||||
| 125 | Effect of antipsychotic drugs on human liver cytochrome P-450 (CYP) isoforms in vitro: preferential inhibition of CYP2D6. Drug Metab Dispos. 1999 Sep;27(9):1078-84. | ||||
| 126 | Identification of human cytochrome p450 isozymes involved in diphenhydramine N-demethylation. Drug Metab Dispos. 2007 Jan;35(1):72-8. | ||||
| 127 | Reappraisal of the role of dolasetron in prevention and treatment of nausea and vomiting associated with surgery or chemotherapy. Cancer Manag Res. 2012;4:67-73. | ||||
| 128 | Characterization of the cytochrome P450 enzymes involved in the in vitro metabolism of ethosuximide by human hepatic microsomal enzymes Xenobiotica. 2003 Mar;33(3):265-76. doi: 10.1080/0049825021000061606. | ||||
| 129 | CYP4F enzymes are responsible for the elimination of fingolimod (FTY720), a novel treatment of relapsing multiple sclerosis. Drug Metab Dispos. 2011 Feb;39(2):191-8. | ||||
| 130 | In vitro identification of the human cytochrome p450 enzymes involved in the oxidative metabolism of loxapine. Biopharm Drug Dispos. 2011 Oct;32(7):398-407. | ||||
| 131 | Cytochrome P450 2D6.1 and cytochrome P450 2D6.10 differ in catalytic activity for multiple substrates. Pharmacogenetics. 2001 Aug;11(6):477-87. | ||||
| 132 | A review of the pharmacological and clinical profile of mirtazapine. CNS Drug Rev. 2001 Fall;7(3):249-64. | ||||
| 133 | A dual system platform for drug metabolism: Nalbuphine as a model compound | ||||
| 134 | Quantitative contribution of CYP2D6 and CYP3A to oxycodone metabolism in human liver and intestinal microsomes. Drug Metab Dispos. 2004 Apr;32(4):447-54. | ||||
| 135 | Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015 Aug;98(2):127-34. | ||||
| 136 | Pazopanib: the newest tyrosine kinase inhibitor for the treatment of advanced or metastatic renal cell carcinoma. Drugs. 2011 Mar 5;71(4):443-54. | ||||
| 137 | The influence of the CYP3A4*22 polymorphism and CYP2D6 polymorphisms on serum concentrations of aripiprazole, haloperidol, pimozide, and risperidone in psychiatric patients. J Clin Psychopharmacol. 2015 Jun;35(3):228-36. | ||||
| 138 | Novel pathways of ponatinib disposition catalyzed by CYP1A1 involving generation of potentially toxic metabolites. J Pharmacol Exp Ther. 2017 Oct;363(1):12-19. | ||||
| 139 | ABCB1 and ABCG2, but not CYP3A4 limit oral availability and brain accumulation of the RET inhibitor pralsetinib | ||||
| 140 | DrugBank(Pharmacology-Metabolism):Pralsetinib | ||||
| 141 | Potential for drug interactions involving cytochromes P450 2D6 and 3A4 on general adult psychiatric and functional elderly psychiatric wards. Br J Clin Pharmacol. 2004 Apr;57(4):464-72. | ||||
| 142 | Effects of propofol on human hepatic microsomal cytochrome P450 activities. Xenobiotica. 1998 Sep;28(9):845-53. | ||||
| 143 | A liquid chromatographic-electrospray-tandem mass spectrometric method for quantitation of quetiapine in human plasma and liver microsomes: application to study in vitro metabolism. J Anal Toxicol. 2004 Sep;28(6):443-8. | ||||
| 144 | Reversible coma caused by risperidone-ritonavir interaction. Clin Neuropharmacol. 2002 Sep-Oct;25(5):251-3. | ||||
| 145 | Clinical pharmacokinetics and pharmacodynamics of solifenacin. Clin Pharmacokinet. 2009;48(5):281-302. | ||||
| 146 | Metabolism of sumatriptan revisited | ||||
| 147 | Identification of CYP3A4 as the primary cytochrome P450 responsible for the metabolism of tandospirone by human liver microsomes | ||||
| 148 | Clinical assessment of drug-drug interactions of tasimelteon, a novel dual melatonin receptor agonist J Clin Pharmacol. 2015 Sep;55(9):1004-11. doi: 10.1002/jcph.507. | ||||
| 149 | Characterization of human cytochromes P450 involved in theophylline 8-hydroxylation. Biochem Pharmacol. 1995 Jul 17;50(2):205-11. | ||||
| 150 | Thioridazine steady-state plasma concentrations are influenced by tobacco smoking and CYP2D6, but not by the CYP2C9 genotype. Eur J Clin Pharmacol. 2003 May;59(1):45-50. | ||||
| 151 | Clinical pharmacology of tramadol. Clin Pharmacokinet. 2004;43(13):879-923. | ||||
| 152 | Cytochrome P450-mediated metabolism of triclosan attenuates its cytotoxicity in hepatic cells | ||||
| 153 | The associations between CYP2D6 metabolizer status and pharmacokinetics and clinical outcomes of venlafaxine: a systematic review and meta-analysis. Pharmacopsychiatry. 2019 Sep;52(5):222-231. | ||||
| 154 | Vilazodone HCl (Viibryd): A Serotonin Partial Agonist and Reuptake Inhibitor For the Treatment of Major Depressive Disorder. P T. 2012 Jan;37(1):28-31. | ||||
| 155 | Identification of the cytochrome P450 and other enzymes involved in the in vitro oxidative metabolism of a novel antidepressant, Lu AA21004. Drug Metab Dispos. 2012 Jul;40(7):1357-65. | ||||
| 156 | Metabolic interactions between acetaminophen (paracetamol) and two flavonoids, luteolin and quercetin, through in-vitro inhibition studies. J Pharm Pharmacol. 2017 Dec;69(12):1762-1772. | ||||
| 157 | Involvement of CYP2D6 in the in vitro metabolism of amphetamine, two N-alkylamphetamines and their 4-methoxylated derivatives. Xenobiotica. 1999 Jul;29(7):719-32. | ||||
| 158 | Aripiprazole: a review of its use in schizophrenia and schizoaffective disorder. Drugs. 2004;64(15):1715-36. | ||||
| 159 | More methemoglobin is produced by benzocaine treatment than lidocaine treatment in human in vitro systems | ||||
| 160 | Binimetinib Is a Potent Reversible and Time-Dependent Inhibitor of Cytochrome P450 1A2 Chem Res Toxicol. 2021 Apr 19;34(4):1169-1174. doi: 10.1021/acs.chemrestox.1c00036. | ||||
| 161 | The roles of CYP2D6 and stereoselectivity in the clinical pharmacokinetics of chlorpheniramine. Br J Clin Pharmacol. 2002 May;53(5):519-25. | ||||
| 162 | Identification of enzymes involved in phase I metabolism of ciclesonide by human liver microsomes. Eur J Drug Metab Pharmacokinet. 2005 Oct-Dec;30(4):275-86. | ||||
| 163 | Clinical pharmacokinetics and pharmacodynamics of dapagliflozin, a selective inhibitor of sodium-glucose co-transporter type 2. Clin Pharmacokinet. 2014 Jan;53(1):17-27. | ||||
| 164 | A long-standing mystery solved: the formation of 3-hydroxydesloratadine is catalyzed by CYP2C8 but prior glucuronidation of desloratadine by UDP-glucuronosyltransferase 2B10 is an obligatory requirement Drug Metab Dispos. 2015 Apr;43(4):523-33. doi: 10.1124/dmd.114.062620. | ||||
| 165 | Effects of the moderate CYP3A4 inhibitor, fluconazole, on the pharmacokinetics of fesoterodine in healthy subjects. Br J Clin Pharmacol. 2011 Aug;72(2):263-9. | ||||
| 166 | Pharmacokinetic drug interactions of gefitinib with rifampicin, itraconazole and metoprolol. Clin Pharmacokinet. 2005;44(10):1067-81. | ||||
| 167 | Influences of CYP2D6*10 polymorphisms on the pharmacokinetics of iloperidone and its metabolites in Chinese patients with schizophrenia: a population pharmacokinetic analysis. Acta Pharmacol Sin. 2016 Nov;37(11):1499-1508. | ||||
| 168 | Exploration of catalytic properties of CYP2D6 and CYP3A4 through metabolic studies of levorphanol and levallorphan | ||||
| 169 | N-demethylation of levo-alpha-acetylmethadol by human placental aromatase. Biochem Pharmacol. 2004 Mar 1;67(5):885-92. | ||||
| 170 | The Role of Levomilnacipran in the Management of Major Depressive Disorder: A Comprehensive Review Curr Neuropharmacol. 2016;14(2):191-9. doi: 10.2174/1570159x14666151117122458. | ||||
| 171 | Identification of cytochrome P450 isoforms involved in the metabolism of loperamide in human liver microsomes. Eur J Clin Pharmacol. 2004 Oct;60(8):575-81. | ||||
| 172 | Pharmacogenomics of statins: understanding susceptibility to adverse effects. Pharmgenomics Pers Med. 2016 Oct 3;9:97-106. | ||||
| 173 | Cytochrome P450 enzymes contributing to demethylation of maprotiline in man. Pharmacol Toxicol. 2002 Mar;90(3):144-9. | ||||
| 174 | Meclizine metabolism and pharmacokinetics: formulation on its absorption. J Clin Pharmacol. 2012 Sep;52(9):1343-9. | ||||
| 175 | Methadone metabolism and drug-drug interactions: in vitro and in vivo literature review. J Pharm Sci. 2018 Dec;107(12):2983-2991. | ||||
| 176 | Lack of interaction between amiodarone and mexiletine in cardiac arrhythmia patients. J Clin Pharmacol. 2002 Mar;42(3):342-6. | ||||
| 177 | Impact of CYP2D6*10 on mexiletine pharmacokinetics in healthy adult volunteers. Eur J Clin Pharmacol. 2003 Sep;59(5-6):395-9. | ||||
| 178 | Akathisia with combined use of midodrine and promethazine. JAMA. 2006 May 3;295(17):2000-1. Letter | ||||
| 179 | Ema.europa:Pitolisant | ||||
| 180 | Involvement of CYP2D6 activity in the N-oxidation of procainamide in man. Pharmacogenetics. 1999 Dec;9(6):683-96. | ||||
| 181 | Oxidation of ranitidine by isozymes of flavin-containing monooxygenase and cytochrome P450. Jpn J Pharmacol. 2000 Oct;84(2):213-20. | ||||
| 182 | PubChem:Tapinarof | ||||
| 183 | Cardiac safety of ophthalmic timolol. Expert Opin Drug Saf. 2016 Nov;15(11):1549-1561. | ||||
| 184 | Effect of the CYP2D6*10 genotype on tolterodine pharmacokinetics. Drug Metab Dispos. 2010 Sep;38(9):1456-63. | ||||
| 185 | Metabolism and pharmacokinetics of the combination Zidovudine plus Lamivudine in the adult Erythrocebus patas monkey determined by liquid chromatography-tandem mass spectrometric analysis | ||||
| 186 | An update on the clinical pharmacology of the dipeptidyl peptidase 4 inhibitor alogliptin used for the treatment of type 2 diabetes mellitus. Clin Exp Pharmacol Physiol. 2015 Dec;42(12):1225-38. | ||||
| 187 | A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab Dispos. 2007 Oct;35(10):1797-805. | ||||
| 188 | Drug Interactions with Angiotensin Receptor Blockers: Role of Human Cytochromes P450 | ||||
| 189 | Lithium Intoxication in the Elderly: A Possible Interaction between Azilsartan, Fluvoxamine, and Lithium | ||||
| 190 | Cariprazine: chemistry, pharmacodynamics, pharmacokinetics, and metabolism, clinical efficacy, safety, and tolerability. Expert Opin Drug Metab Toxicol. 2013 Feb;9(2):193-206. | ||||
| 191 | Involvement of CYP2D1 in the metabolism of carteolol by male rat liver microsomes. Xenobiotica. 1997 Nov;27(11):1121-9. | ||||
| 192 | Celecoxib is a substrate of CYP2D6: impact on celecoxib metabolism in individuals with CYP2C9*3 variants. Drug Metab Pharmacokinet. 2018 Oct;33(5):219-227. | ||||
| 193 | Effects of CYP3A inhibition on the metabolism of cilostazol. Clin Pharmacokinet. 1999;37 Suppl 2:61-8. | ||||
| 194 | Oxidative metabolism of flunarizine and cinnarizine by microsomes from B-lymphoblastoid cell lines expressing human cytochrome P450 enzymes. Biol Pharm Bull. 1996 Nov;19(11):1511-4. | ||||
| 195 | Eletriptan metabolism by human hepatic CYP450 enzymes and transport by human P-glycoprotein. Drug Metab Dispos. 2003 Jul;31(7):861-9. | ||||
| 196 | CYP2D6 P34S Polymorphism and Outcomes of Escitalopram Treatment in Koreans with Major Depression. Psychiatry Investig. 2013 Sep;10(3):286-93. | ||||
| 197 | Flecainide: current status and perspectives in arrhythmia management. World J Cardiol. 2015 Feb 26;7(2):76-85. | ||||
| 198 | LABEL:DDYI- flibanserin tablet, film coated | ||||
| 199 | Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A Genotypes and Serotonin Reuptake Inhibitor Antidepressants | ||||
| 200 | Effect of cytochrome P450 2D1 inhibition on hydrocodone metabolism and its behavioral consequences in rats. J Pharmacol Exp Ther. 1997 Mar;280(3):1374-82. | ||||
| 201 | An updated review of iclaprim: a potent and rapidly bactericidal antibiotic for the treatment of skin and skin structure infections and nosocomial pneumonia caused by gram-positive including multidrug-resistant bacteria. Open Forum Infect Dis. 2018 Jan 6;5(2):ofy003. | ||||
| 202 | In vitro characterization of the inhibition profile of loratadine, desloratadine, and 3-OH-desloratadine for five human cytochrome P-450 enzymes. Drug Metab Dispos. 2001 Sep;29(9):1173-5. | ||||
| 203 | Ethnic differences in genetic polymorphisms of CYP2D6, CYP2C19, CYP3As and MDR1/ABCB1. Drug Metab Pharmacokinet. 2004 Apr;19(2):83-95. | ||||
| 204 | Inhibitory effects of nicardipine to cytochrome P450 (CYP) in human liver microsomes. Biol Pharm Bull. 2005 May;28(5):882-5. | ||||
| 205 | Molecular basis of polymorphic drug metabolism. J Mol Med (Berl). 1995 Nov;73(11):539-53. | ||||
| 206 | Inhibitory effects of antiparasitic drugs on cytochrome P450 2D6. Eur J Clin Pharmacol. 1995;48(1):35-8. | ||||
| 207 | Interpreting serum risperidone concentrations. Pharmacotherapy. 2005 Feb;25(2):299-302. | ||||
| 208 | DrugBank(Pharmacology-Metabolism):Phenmetrazine | ||||
| 209 | Physiologically Based Pharmacokinetic Modeling of Transdermal Selegiline and Its Metabolites for the Evaluation of Disposition Differences between Healthy and Special Populations | ||||
| 210 | Inter- and intraindividual pharmacokinetic variations in the treatment of Parkinson's disease. Rinsho Shinkeigaku. 2005 Nov;45(11):895-8. | ||||
| 211 | Some aspects of genetic polymorphism in the biotransformation of antidepressants. Therapie. 2004 Jan-Feb;59(1):5-12. | ||||
| 212 | Role of tetrabenazine for Huntington's disease-associated chorea. Ann Pharmacother. 2010 Jun;44(6):1080-9. | ||||
| 213 | Tiotropium bromide. A review of its use as maintenance therapy in patients with COPD. Treat Respir Med. 2004;3(4):247-68. | ||||
| 214 | Tiotropium rromide/olodaterol (Stiolto Respimat): once-daily combination therapy for the maintenance of COPD. P T. 2016 Feb;41(2):97-102. | ||||
| 215 | FDA Label of Egaten. The 2020 official website of the U.S. Food and Drug Administration. | ||||
| 216 | Galactorrhea during treatment with trimipramine. A case report. Pharmacopsychiatry. 2005 Nov;38(6):326-7. | ||||
| 217 | Pharmacokinetics, toxicological and clinical aspects of ulipristal acetate: insights into the mechanisms implicated in the hepatic toxicity Drug Metab Rev. 2021 Aug;53(3):375-383. doi: 10.1080/03602532.2021.1917599. | ||||
| 218 | Pharmacokinetics, metabolite profiling, safety, and tolerability of inhalation aerosol of 101BHG-D01, a novel, long-acting and selective muscarinic receptor antagonist, in healthy Chinese subjects | ||||
| 219 | DrugBank(Pharmacology-Metabolism):Umeclidinium | ||||
| 220 | Tardive dyskinesia: placing vesicular monoamine transporter type 2 (VMAT2) inhibitors into clinical perspective. Expert Rev Neurother. 2018 Apr;18(4):323-332. | ||||
| 221 | Mechanism of cytochrome P4503A4- and 2D6-catalyzed dehydrogenation of ezlopitant as probed with isotope effects using five deuterated analogs. Drug Metab Dispos. 2001 Dec;29(12):1599-607. | ||||
| 222 | Drug-drug interactions of beta-adrenoceptor blockers. Arzneimittelforschung. 2003;53(12):814-22. | ||||
| 223 | Halofantrine and chloroquine inhibit CYP2D6 activity in healthy Zambians. Br J Clin Pharmacol. 1998 Mar;45(3):315-7. | ||||
| 224 | Metabolism and Interactions of Chloroquine and Hydroxychloroquine with Human Cytochrome P450 Enzymes and Drug Transporters Curr Drug Metab. 2020;21(14):1127-1135. doi: 10.2174/1389200221999201208211537. | ||||
| 225 | Prediction of human liver microsomal oxidations of 7-ethoxycoumarin and chlorzoxazone with kinetic parameters of recombinant cytochrome P-450 enzymes. Drug Metab Dispos. 1999 Nov;27(11):1274-80. | ||||
| 226 | Human cytochrome p450 induction and inhibition potential of clevidipine and its primary metabolite h152/81. Drug Metab Dispos. 2006 May;34(5):734-7. | ||||
| 227 | Phase 1 study to investigate the pharmacokinetic properties of dacomitinib in healthy adult Chinese subjects genotyped for CYP2D6. Xenobiotica. 2018 May;48(5):459-466. | ||||
| 228 | Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51(1):73-8. | ||||
| 229 | Development of encorafenib for BRAF-mutated advanced melanoma. Curr Opin Oncol. 2018 Mar;30(2):125-133. | ||||
| 230 | Metabolism of epinastine, a histamine H1 receptor antagonist, in human liver microsomes in comparison with that of terfenadine. Res Commun Mol Pathol Pharmacol. 1997 Dec;98(3):273-92. | ||||
| 231 | Pharmacogenetic Gene-Drug Associations in Pediatric Burn and Surgery Patients | ||||
| 232 | Engineering of cytochrome P450 3A4 for enhanced peroxide-mediated substrate oxidation using directed evolution and site-directed mutagenesis. Drug Metab Dispos. 2006 Dec;34(12):1958-65. | ||||
| 233 | Cross-species comparisons of the pharmacokinetics of ibudilast | ||||
| 234 | In vitro evaluation of cytochrome P450-mediated drug interactions between cytarabine, idarubicin, itraconazole and caspofungin. Hematology. 2004 Jun;9(3):217-21. | ||||
| 235 | Drugs and steatohepatitis. Semin Liver Dis. 2002;22(2):185-94. | ||||
| 236 | Nebivolol: a review of its clinical and pharmacological characteristics. Int J Clin Pharmacol Ther. 2006 Aug;44(8):344-57. | ||||
| 237 | A study on CYP2C19 and CYP2D6 polymorphic effects on pharmacokinetics and pharmacodynamics of amitriptyline in healthy Koreans. Clin Transl Sci. 2017 Mar;10(2):93-101. | ||||
| 238 | Polymorphic cytochrome P450 2D6: humanized mouse model and endogenous substrates | ||||
| 239 | Chemistry, pharmacology, toxicology, and hepatic metabolism of designer drugs of the amphetamine (ecstasy), piperazine, and pyrrolidinophenone types: a synopsis. Ther Drug Monit. 2004 Apr;26(2):127-31. | ||||
| 240 | Crystal structure of human cytochrome P450 2D6 with prinomastat bound | ||||
| 241 | Xenobiotic-metabolizing cytochromes p450 in human white adipose tissue: expression and induction. Drug Metab Dispos. 2010 Apr;38(4):679-86. | ||||
| 242 | DrugBank(Pharmacology-Metabolism):Ripretinib | ||||
| 243 | Cytochrome P450 3A inhibitor itraconazole affects plasma concentrations of risperidone and 9-hydroxyrisperidone in schizophrenic patients. Clin Pharmacol Ther. 2005 Nov;78(5):520-8. | ||||
| 244 | SULT 1A3 single-nucleotide polymorphism and the single dose pharmacokinetics of inhaled salbutamol enantiomers: are some athletes at risk of higher urine levels? Drug Test Anal. 2015 Feb;7(2):109-13. doi: 10.1002/dta.1645. | ||||
| 245 | Tafenoquine and its potential in the treatment and relapse prevention of Plasmodium vivax malaria: the evidence to date. Drug Des Devel Ther. 2016 Jul 26;10:2387-99. | ||||
| 246 | In vitro metabolism of the opioid tilidine and interaction of tilidine and nortilidine with CYP3A4, CYP2C19, and CYP2D6 | ||||
| 247 | Protease inhibitors in patients with HIV disease. Clinically important pharmacokinetic considerations. Clin Pharmacokinet. 1997 Mar;32(3):194-209. | ||||
| 248 | Serum flecainide S/R ratio reflects the CYP2D6 genotype and changes in CYP2D6 activity. Drug Metab Pharmacokinet. 2015 Aug;30(4):257-62. | ||||
| 249 | Population pharmacokinetic analysis of mirtazapine. Eur J Clin Pharmacol. 2004 Sep;60(7):473-80. | ||||
| 250 | DrugBank(Pharmacology-Metabolism):Elagolix sodium | ||||
| 251 | Identification of cytochrome P450 2E1 as the predominant enzyme catalyzing human liver microsomal defluorination of sevoflurane, isoflurane, and methoxyflurane. Anesthesiology. 1993 Oct;79(4):795-807. | ||||
| 252 | Therapeutic Drug Monitoring of Rivastigmine and Donepezil Under Consideration of CYP2D6 Genotype-Dependent Metabolism of Donepezil | ||||
| 253 | Population pharmacokinetics of upadacitinib using the immediate-release and extended-release formulations in healthy subjects and subjects with rheumatoid arthritis: analyses of phase I-III clinical trials. Clin Pharmacokinet. 2019 Aug;58(8):1045-1058. | ||||
| 254 | PubChem:Upadacitinib | ||||
| 255 | CYP2D plays a major role in berberine metabolism in liver of mice and humans | ||||
| 256 | Metabolic drug interactions with newer antipsychotics: a comparative review. Basic Clin Pharmacol Toxicol. 2007 Jan;100(1):4-22. | ||||
| 257 | Comparative molecular field analysis and QSAR on substrates binding to cytochrome p450 2D6. Bioorg Med Chem. 2003 Dec 1;11(24):5545-54. | ||||
| 258 | Dapoxetine: a new option in the medical management of premature ejaculation. Ther Adv Urol. 2012 Oct;4(5):233-51. | ||||
| 259 | No influence of the CYP2C19-selective inhibitor omeprazole on the pharmacokinetics of the dopamine receptor agonist rotigotine | ||||
| 260 | Stereoselective metabolism of cibenzoline, an antiarrhythmic drug, by human and rat liver microsomes: possible involvement of CYP2D and CYP3A. Drug Metab Dispos. 2000 Sep;28(9):1128-34. | ||||
| 261 | Assessment of CYP2D6 activity as a form of optimizing antidepressant therapy. Psychiatr Pol. 2004 Nov-Dec;38(6):1093-104. | ||||
| 262 | In vitro metabolism of TAK-438, vonoprazan fumarate, a novel potassium-competitive acid blocker. Xenobiotica. 2017 Dec;47(12):1027-1034. | ||||
| 263 | Assessments of CYP?inhibition?based drug-drug interaction between vonoprazan and poziotinib in?vitro and in?vivo | ||||
| 264 | Identification of cytochrome P450 enzymes involved in the metabolism of zotepine, an antipsychotic drug, in human liver microsomes. Xenobiotica. 1999 Mar;29(3):217-29. | ||||
| 265 | Involvement of multiple human cytochromes P450 in the liver microsomal metabolism of astemizole and a comparison with terfenadine. Br J Clin Pharmacol. 2001 Feb;51(2):133-42. | ||||
| 266 | Metabolism of dexfenfluramine in human liver microsomes and by recombinant enzymes: role of CYP2D6 and 1A2. Pharmacogenetics. 1998 Oct;8(5):423-32. | ||||
| 267 | The role of active metabolites in dihydrocodeine effects. Int J Clin Pharmacol Ther. 2003 Mar;41(3):95-106. | ||||
| 268 | Pharmacokinetic evaluation of idebenone. Expert Opin Drug Metab Toxicol. 2010 Nov;6(11):1437-44. | ||||
| 269 | Differences in the In Vivo and In Vitro Metabolism of Imrecoxib in Humans: Formation of the Rate-Limiting Aldehyde Intermediate Drug Metab Dispos. 2018 Sep;46(9):1320-1328. doi: 10.1124/dmd.118.081182. | ||||
| 270 | Medical Toxicology of Drug Abuse: Synthesized Chemicals and Psychoactive Plants. doi: 10.1002/9781118105955.ch13 | ||||
| 271 | Human cytochromes P450 mediating phenacetin O-deethylation in vitro: validation of the high affinity component as an index of CYP1A2 activity. J Pharm Sci. 1998 Dec;87(12):1502-7. | ||||
| 272 | Genotype and co-medication dependent CYP2D6 metabolic activity: effects on serum concentrations of aripiprazole, haloperidol, risperidone, paliperidone and zuclopenthixol. Eur J Clin Pharmacol. 2016 Feb;72(2):175-84. | ||||
| 273 | Contribution of human hepatic cytochrome P450s and steroidogenic CYP17 to the N-demethylation of aminopyrine. Xenobiotica. 1999 Feb;29(2):187-93. | ||||
| 274 | Different effects of inhibitors on the O- and N-demethylation of codeine in human liver microsomes. Eur J Clin Pharmacol. 1997;52(1):41-7. | ||||
| 275 | Effects of the antifungal agents on oxidative drug metabolism: clinical relevance. Clin Pharmacokinet. 2000 Feb;38(2):111-80. | ||||
| 276 | Cytochrome P-450 enzymes and FMO3 contribute to the disposition of the antipsychotic drug perazine in vitro. Psychopharmacology (Berl). 2000 Sep;151(4):312-20. | ||||
| 277 | DMW/ISSX 2000 Meeting. The Drug Metabolism Workshop/International Society for the Study of Xenobiotics. St. Andrews, Scotland, June 11-16, 2000. Abstracts. Drug Metab Rev. 2000;32 Suppl 1:1-136. | ||||
| 278 | CYP2D6 is primarily responsible for the metabolism of clomiphene. Drug Metab Pharmacokinet. 2008;23(2):101-5. | ||||
| 279 | VMAT2 Inhibitors in Neuropsychiatric Disorders | ||||
| 280 | DrugBank(Pharmacology-Metabolism)Lofexidine | ||||
| 281 | Excretion and metabolism of milnacipran in humans after oral administration of milnacipran hydrochloride Drug Metab Dispos. 2012 Sep;40(9):1723-35. doi: 10.1124/dmd.112.045120. | ||||
| 282 | FDA LABEL:Milnacipran | ||||
| 283 | Serotonin and Norepinephrine Reuptake Inhibitors | ||||
| 284 | Management of postoperative nausea and vomiting: focus on palonosetron | ||||
| 285 | Phase I pharmacokinetic study of the novel antitumor agent SR233377. Clin Cancer Res. 2000 Aug;6(8):3088-94. | ||||
| 286 | Residual effects of esmirtazapine on actual driving performance: overall findings and an exploratory analysis into the role of CYP2D6 phenotype. Psychopharmacology (Berl). 2011 May;215(2):321-32. | ||||
| 287 | Involvement of CYP2D6 but not CYP2C19 in nicergoline metabolism in humans. Br J Clin Pharmacol. 1996 Dec;42(6):707-11. | ||||
| 288 | Cytochrome P450 2D catalyze steroid 21-hydroxylation in the brain. Endocrinology. 2004 Feb;145(2):699-705. | ||||
| 289 | Relationship of cytochrome P450 pharmacogenetics to the effects of yohimbine on gastrointestinal transit and catecholamines in healthy subjects. Neurogastroenterol Motil. 2008 Aug;20(8):891-9. | ||||
| 290 | Plasma protein binding, pharmacokinetics, tissue distribution and CYP450 biotransformation studies of fidarestat by ultra high performance liquid chromatography-high resolution mass spectrometry | ||||
| 291 | Preclinical pharmacokinetics and disposition of a novel selective VEGFR inhibitor fruquintinib (HMPL-013) and the prediction of its human pharmacokinetics | ||||
| 292 | Stable expression of human cytochrome P450 3A4 in V79 cells and its application for metabolic profiling of ergot derivatives. Eur J Pharmacol. 1995 Oct 6;293(3):183-90. | ||||
| 293 | Involvement of multiple cytochrome P450 and UDP-glucuronosyltransferase enzymes in the in vitro metabolism of muraglitazar. Drug Metab Dispos. 2007 Jan;35(1):139-49. | ||||
| 294 | Asunaprevir: a review of preclinical and clinical pharmacokinetics and drug-drug interactions. Clin Pharmacokinet. 2015 Dec;54(12):1205-22. | ||||
| 295 | Pharmacokinetics, Disposition, and Biotransformation of [(14)C]Omecamtiv Mecarbil in Healthy Male Subjects after a Single Intravenous or Oral Dose | ||||
| 296 | Metabolism and disposition of hepatitis C polymerase inhibitor dasabuvir in humans. Drug Metab Dispos. 2016 Aug;44(8):1139-47. | ||||
| 297 | DrugBank(Pharmacology-Metabolism):Chlorpromazine | ||||
| 298 | In vitro metabolism of the analgesic bicifadine in the mouse, rat, monkey, and human. Drug Metab Dispos. 2007 Dec;35(12):2232-41. | ||||
| 299 | Characterization of human cytochrome P450 enzymes catalyzing domperidone N-dealkylation and hydroxylation in vitro. Br J Clin Pharmacol. 2004 Sep;58(3):277-87. | ||||
| 300 | The metabolic profile of azimilide in man: in vivo and in vitro evaluations. J Pharm Sci. 2005 Sep;94(9):2084-95. | ||||
| 301 | Moclobemide, a substrate of CYP2C19 and an inhibitor of CYP2C19, CYP2D6, and CYP1A2: a panel study. Clin Pharmacol Ther. 1995 Jun;57(6):670-7. | ||||
| 302 | The relative contribution of monoamine oxidase and cytochrome p450 isozymes to the metabolic deamination of the trace amine tryptamine | ||||
| 303 | Metabolism and disposition of the SGLT2 inhibitor bexagliflozin in rats, monkeys and humans | ||||
| 304 | The potent mechanism-based inactivation of CYP2D6 and CYP3A4 with fusidic acid in in vivo bioaccumulation. Xenobiotica. 2018 Oct;48(10):999-1005. | ||||
| 305 | DrugBank(Pharmacology-Metabolism):BMS-986165 | ||||
| 306 | Idalopirdine as a treatment for Alzheimer's disease. Expert Opin Investig Drugs. 2015;24(7):981-7. | ||||
| 307 | Disposition and metabolism of semagacestat, a {gamma}-secretase inhibitor, in humans. Drug Metab Dispos. 2010 Apr;38(4):554-65. | ||||
| 308 | Effect of netupitant, a highly selective NK?receptor antagonist, on the pharmacokinetics of palonosetron and impact of the fixed dose combination of netupitant and palonosetron when coadministered with ketoconazole, rifampicin, and oral contraceptives. Support Care Cancer. 2013 Oct;21(10):2879-87. | ||||
| 309 | CYP2B6, CYP2D6, and CYP3A4 catalyze the primary oxidative metabolism of perhexiline enantiomers by human liver microsomes. Drug Metab Dispos. 2007 Jan;35(1):128-38. | ||||
| 310 | A Phase I Study of ABC294640, a First-in-Class Sphingosine Kinase-2 Inhibitor, in Patients with Advanced Solid Tumors | ||||
| 311 | DrugBank(Pharmacology-Metabolism)BRN-1907611 | ||||
| 312 | Characterization of human cytochrome P450 enzymes involved in the biotransformation of eperisone. Xenobiotica. 2009 Jan;39(1):1-10. | ||||
| 313 | In vitro evaluation of fenfluramine and norfenfluramine as victims of drug interactions | ||||
| 314 | Plasma pharmacokinetics and CYP3A12-dependent metabolism of c-kit inhibitor imatinib in dogs | ||||
| 315 | Pharmacodynamic and pharmacokinetic behavior of landiolol during dobutamine challenge in healthy adults | ||||
| 316 | Pharmacokinetics of novel atrial-selective antiarrhythmic agent vernakalant hydrochloride injection (RSD1235): influence of CYP2D6 expression and other factors. J Clin Pharmacol. 2009 Jan;49(1):17-29. | ||||
| 317 | Evaluation of in vitro drug interactions with karenitecin, a novel, highly lipophilic camptothecin derivative in phase II clinical development. J Clin Pharmacol. 2003 Sep;43(9):1008-14. | ||||
| 318 | DrugBank(Pharmacology-Metabolism):Ganaxolone | ||||
| 319 | DrugBank(Pharmacology-Metabolism):MRTX849 | ||||
| 320 | Nefazodone pharmacokinetics in depressed children and adolescents. J Am Acad Child Adolesc Psychiatry. 2000 Aug;39(8):1008-16. | ||||
| 321 | In Vitro ADME and Preclinical Pharmacokinetics of Ulotaront, a TAAR1/5-HT(1A) Receptor Agonist for the Treatment of Schizophrenia | ||||
| 322 | In vitro biotransformation of sildenafil (Viagra): identification of human cytochromes and potential drug interactions. Drug Metab Dispos. 2000 Apr;28(4):392-7. | ||||
| 323 | DrugBank(Pharmacology-Metabolism):Ag-221 | ||||
| 324 | Effect of low-dose ritonavir on the pharmacokinetics of the CXCR4 antagonist AMD070 in healthy volunteers | ||||
| 325 | Quantitative Assessment of Elagolix Enzyme-Transporter Interplay and Drug-Drug Interactions Using Physiologically Based Pharmacokinetic Modeling | ||||
| 326 | Interpretation of opiate urine drug screens. | ||||
| 327 | Oxidative Deamination of Emixustat by Human Vascular Adhesion Protein-1/Semicarbazide-Sensitive Amine Oxidase | ||||
| 328 | DrugBank(Pharmacology-Metabolism):Verapamil | ||||
| 329 | Novel binding mode of the acidic CYP2D6 substrates pactimibe and its metabolite R-125528. Drug Metab Dispos. 2008 Sep;36(9):1938-43. | ||||
| 330 | Determination of an optimal dosing regimen for fexinidazole, a novel oral drug for the treatment of human African trypanosomiasis: first-in-human studies | ||||
| 331 | Fexinidazole--a new oral nitroimidazole drug candidate entering clinical development for the treatment of sleeping sickness. PLoS Negl Trop Dis. 2010 Dec 21;4(12):e923. | ||||
| 332 | Metabolism, excretion, and pharmacokinetics of ((3,3-difluoropyrrolidin-1-yl)((2S,4S)-4-(4-(pyrimidin-2-yl)piperazin-1-yl)pyrrolidin-2-yl)methanone, a dipeptidyl peptidase inhibitor, in rat, dog and human | ||||
| 333 | CYP3A but not P-gp plays a relevant role in the in vivo intestinal and hepatic clearance of the delta-specific phosphoinositide-3 kinase inhibitor leniolisib | ||||
| 334 | Metabolic disposition of AZD8931, an oral equipotent inhibitor of EGFR, HER2 and HER3 signalling, in rat, dog and man | ||||
| 335 | Phenotypic and metabolic investigation of a CSF-1R kinase receptor inhibitor (BLZ945) and its pharmacologically active metabolite | ||||
| 336 | Metabolism and disposition of [14C]BMS-690514 after oral administration to rats, rabbits, and dogs | ||||
| 337 | Functional analysis of CYP2D6.31 variant: homology modeling suggests possible disruption of redox partner interaction by Arg440His substitution. Proteins. 2005 May 1;59(2):339-46. | ||||
| 338 | Human pharmacology of MDMA: pharmacokinetics, metabolism, and disposition. Ther Drug Monit. 2004 Apr;26(2):137-44. | ||||
| 339 | Cytochrome P450 enzymes involved in the metabolic pathway of the histamine 2 (H2)-receptor antagonist roxatidine acetate by human liver microsomes. Arzneimittelforschung. 2001;51(8):651-8. | ||||
| 340 | Interspecies pharmacokinetics and in vitro metabolism of SQ109. Br J Pharmacol. 2006 Mar;147(5):476-85. | ||||
| 341 | Disposition of the antipsychotic agent CI-1007 in rats, monkeys, dogs, and human cytochrome P450 2D6 extensive metabolizers. Species comparison and allometric scaling | ||||
| 342 | Metabolic Profiling of the Novel Hypoxia-Inducible Factor 2 Inhibitor PT2385 In Vivo and In Vitro | ||||
| 343 | Metabolism and disposition of vatalanib (PTK787/ZK-222584) in cancer patients | ||||
| 344 | Vatalanib population pharmacokinetics in patients with myelodysplastic syndrome: CALGB 10105 (Alliance) | ||||
| 345 | Involvement of human cytochrome P450 2D6 in the bioactivation of acetaminophen. Drug Metab Dispos. 2000 Dec;28(12):1397-400. | ||||
| 346 | Metabolism, pharmacokinetics, and excretion of a highly selective N-methyl-D-aspartate receptor antagonist, traxoprodil, in human cytochrome P450 2D6 extensive and poor metabolizers | ||||
| 347 | In vitro metabolism of ferroquine (SSR97193) in animal and human hepatic models and antimalarial activity of major metabolites on Plasmodium falciparum. Drug Metab Dispos. 2006 Apr;34(4):667-82. | ||||
| 348 | Effect of deuteration on metabolism and clearance of Nerispirdine (HP184) and AVE5638 | ||||
| 349 | Metabolism of the newest antidepressants: comparisons with related predecessors. IDrugs. 2004 Feb;7(2):143-50. | ||||
| 350 | Cytochrome P450 Mediated Bioactivation of Saracatinib | ||||
| 351 | Identification of metabolic pathways involved in the biotransformation of tolperisone by human microsomal enzymes. Drug Metab Dispos. 2003 May;31(5):631-6. | ||||
| 352 | In vitro metabolism of CP-122,721 ((2S,3S)-2-phenyl-3-[(5-trifluoromethoxy-2-methoxy)benzylamino]piperidine), a non-peptide antagonist of the substance P receptor. Drug Metab Pharmacokinet. 2007 Oct;22(5):336-49. | ||||
| 353 | Characterization of human cytochrome P450 isoenzymes involved in the metabolism of vinorelbine. Fundam Clin Pharmacol. 2005 Oct;19(5):545-53. | ||||
| 354 | Evaluation of the inhibitory and induction potential of YM758, a novel If channel inhibitor, for human P450-mediated metabolism | ||||
| 355 | Pharmacokinetics, tissue distribution and identification of putative metabolites of JI-101 - a novel triple kinase inhibitor in rats | ||||
| 356 | Disposition and metabolism of LY2603618, a Chk-1 inhibitor following intravenous administration in patients with advanced and/or metastatic solid tumors. Xenobiotica. 2014 Sep;44(9):827-41. | ||||
| 357 | Differential cytochrome P450 2D metabolism alters tafenoquine pharmacokinetics. Antimicrob Agents Chemother. 2015 Jul;59(7):3864-9. | ||||
| 358 | Physiologically based pharmacokinetic modeling to predict complex drug-drug interactions: a case study of AZD2327 and its metabolite, competitive and time-dependent CYP3A inhibitors | ||||
| 359 | Comparative pharmacokinetic profile of cimicoxib in dogs and cats after IV administration | ||||
| 360 | The pharmacogenetics of the new-generation antipsychotics - A scoping review focused on patients with severe psychiatric disorders | ||||
| 361 | In vitro metabolism, pharmacokinetics and drug interaction potentials of emvododstat, a DHODH inhibitor | ||||
| 362 | Evaluation of the drug-drug interaction potential for trazpiroben (TAK-906), a D(2)/D(3) receptor antagonist for gastroparesis, towards cytochrome P450s and transporters | ||||
| 363 | DrugBank(Pharmacology-Metabolism):KD025 | ||||
| 364 | Identification of human P450 isoforms involved in the metabolism of the antiallergic drug, oxatomide, and its kinetic parameters and inhibition constants. Biol Pharm Bull. 2005 Feb;28(2):328-34. | ||||
| 365 | Pharmacokinetics of N-propylajmaline in relation to polymorphic sparteine oxidation. Klin Wochenschr. 1985 Nov 15;63(22):1180-6. | ||||
| 366 | First clinical experience with TRV130: pharmacokinetics and pharmacodynamics in healthy volunteers. J Clin Pharmacol. 2014 Mar;54(3):351-7. | ||||
| 367 | Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48(12):761-804. | ||||
| 368 | Metabolism and pharmacokinetics of allitinib in cancer patients: the roles of cytochrome P450s and epoxide hydrolase in its biotransformation | ||||
| 369 | Pharmacokinetics and Metabolism of Naringin and Active Metabolite Naringenin in Rats, Dogs, Humans, and the Differences Between Species | ||||
| 370 | Nonclinical pharmacokinetics and in vitro metabolism of H3B-6545, a novel selective ERalpha covalent antagonist (SERCA). Cancer Chemother Pharmacol. 2019 Jan;83(1):151-160. | ||||
| 371 | Steady-state pharmacokinetics of a new antipsychotic agent perospirone and its active metabolite, and its relationship with prolactin response. Ther Drug Monit. 2004 Aug;26(4):361-5. | ||||
| 372 | In vitro metabolism of 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin in human liver microsomes | ||||
| 373 | In vitro and in vivo clinical pharmacology of dimethyl benzoylphenylurea, a novel oral tubulin-interactive agent | ||||
| 374 | Phase I study of continuous weekly dosing of dimethylamino benzoylphenylurea (BPU) in patients with solid tumours | ||||
| 375 | Metabolism and disposition of the dipeptidyl peptidase IV inhibitor teneligliptin in humans | ||||
| 376 | Development of a liquid chromatography/tandem-mass spectrometry assay for the simultaneous determination of teneligliptin and its active metabolite teneligliptin sulfoxide in human plasma | ||||
| 377 | Nonlinear mixed effects model analysis of the pharmacokinetics of routinely administered bepridil in Japanese patients with arrhythmias. Biol Pharm Bull. 2006 Mar;29(3):517-21. | ||||
| 378 | Evaluation of the transport, in vitro metabolism and pharmacokinetics of Salvinorin A, a potent hallucinogen. Eur J Pharm Biopharm. 2009 Jun;72(2):471-7. | ||||
| 379 | Tricyclic antidepressant pharmacology and therapeutic drug interactions updated. Br J Pharmacol. 2007 Jul;151(6):737-48. | ||||
| 380 | Inactivation of cytochrome P450 (P450) 3A4 but not P450 3A5 by OSI-930, a thiophene-containing anticancer drug | ||||
| 381 | ITX 5061 quantitation in human plasma with reverse phase liquid chromatography and mass spectrometry detection. Antivir Ther. 2013;18(3):329-36. | ||||
| 382 | Cilomilast: a second generation phosphodiesterase 4 inhibitor for asthma and chronic obstructive pulmonary disease | ||||
| 383 | In vitro metabolism of the novel, highly selective oral angiogenesis inhibitor motesanib diphosphate in preclinical species and in humans | ||||
| 384 | An assessment of human liver-derived in vitro systems to predict the in vivo metabolism and clearance of almokalant | ||||
| 385 | A pharmacokinetic interaction study of avitriptan and propranolol | ||||
| 386 | In vitro metabolism of 7 neuronal nicotinic receptor agonist AZD0328 and enzyme identification for its N-oxide metabolite | ||||
| 387 | Main metabolic pathways of TAK-802, a novel drug candidate for voiding dysfunction, in humans: the involvement of carbonyl reduction by 11-hydroxysteroid dehydrogenase 1 | ||||
| 388 | Identification of cytochrome P450 isoform involved in the metabolism of YM992, a novel selective serotonin re-uptake inhibitor, in human liver microsomes | ||||
| 389 | Behavior of thymidylate kinase toward monophosphate metabolites and its role in the metabolism of 1-(2'-deoxy-2'-fluoro-beta-L-arabinofuranosyl)-5-methyluracil (Clevudine) and 2',3'-didehydro-2',3'-dideoxythymidine in cells. Antimicrob Agents Chemother. 2005 May;49(5):2044-9. doi: 10.1128/AAC.49.5.2044-2049.2005. | ||||
| 390 | Evaluation of the effects of AZD3480 on cardiac repolarization: a thorough QT/QTc study using moxifloxacin as a positive control | ||||
| 391 | Species-dependent metabolism of a novel selective 7 neuronal acetylcholine receptor agonist ABT-107 | ||||
| 392 | In vitro interactions between a potential muscle relaxant E2101 and human cytochromes P450 | ||||
| 393 | In-vitro metabolism of YM17E, an inhibitor of acyl coenzyme A:cholesterol acyltransferase, by liver microsomes in man | ||||
| 394 | Metabolism of ezlopitant, a nonpeptidic substance P receptor antagonist, in liver microsomes: enzyme kinetics, cytochrome P450 isoform identity, and in vitro-in vivo correlation. Drug Metab Dispos. 2000 Sep;28(9):1069-76. | ||||
| 395 | In vitro evaluation of potential in vivo probes for human flavin-containing monooxygenase (FMO): metabolism of benzydamine and caffeine by FMO and P450 isoforms. Br J Clin Pharmacol. 2000 Oct;50(4):311-4. | ||||
| 396 | Pharmacokinetic assessment of the sites of first-pass metabolism of BMS-181101, an antidepressant agent, in rats. J Pharm Pharmacol. 1998 Mar;50(3):275-8. | ||||
| 397 | Pharmacokinetics of sabeluzole and dextromethorphan oxidation capacity in patients with severe hepatic dysfunction and healthy volunteers. Br J Clin Pharmacol. 2001 Feb;51(2):164-8. | ||||
| 398 | Role of cytochrome P4502D6 in the metabolism of brofaromine. A new selective MAO-A inhibitor. Eur J Clin Pharmacol. 1993;45(3):265-9. | ||||
| 399 | Antidepressants: Past, Present and Future. Edited by Sheldon H. Preskorn Christina Y. Stanga John P. Feighner Ruth Ross. Page: 574. | ||||
| 400 | Dopamine formation from tyramine by CYP2D6. Biochem Biophys Res Commun. 1998 Aug 28;249(3):838-43. | ||||
| 401 | Proof of a 1-(3-chlorophenyl)piperazine (mCPP) intake: use as adulterant of cocaine resulting in drug-drug interactions? J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Aug 15;855(2):127-33. | ||||
| 402 | Identification of cytochrome P450 enzymes involved in the metabolism of 4'-methoxy-alpha-pyrrolidinopropiophenone (MOPPP), a designer drug, in human liver microsomes. Xenobiotica. 2003 Oct;33(10):989-98. | ||||
| 403 | N,N-diethyl-2-[4-(phenylmethyl)phenoxy] ethanamine (DPPE) a chemopotentiating and cytoprotective agent in clinical trials: interaction with histamine at cytochrome P450 3A4 and other isozymes that metabolize antineoplastic drugs. Cancer Chemother Pharmacol. 2000;45(4):298-304. | ||||
| 404 | Using chimeric mice with humanized livers to predict human drug metabolism and a drug-drug interaction. J Pharmacol Exp Ther. 2013 Feb;344(2):388-96. | ||||
| 405 | Metabolism of ipecac alkaloids cephaeline and emetine by human hepatic microsomal cytochrome P450s, and their inhibitory effects on P450 enzyme activities. Biol Pharm Bull. 2001 Jun;24(6):678-82. | ||||
| 406 | A sensitive LC-MS/MS assay for the determination of dextromethorphan and metabolites in human urine--application for drug interaction studies assessing potential CYP3A and CYP2D6 inhibition. J Pharm Biomed Anal. 2002 Aug 22;30(1):113-24. | ||||
| 407 | Cytochrome b5 is a major determinant of human cytochrome P450 CYP2D6 and CYP3A4 activity in vivo. Mol Pharmacol. 2015 Apr;87(4):733-9. | ||||
| 408 | Studies on the metabolism of the fentanyl-derived designer drug butyrfentanyl in human in vitro liver preparations and authentic human samples using liquid chromatography-high resolution mass spectrometry (LC-HRMS). Drug Test Anal. 2017 Jul;9(7):1085-1092. | ||||
| 409 | The effect of itraconazole on the pharmacokinetics and pharmacodynamics of bromazepam in healthy volunteers | ||||
| 410 | Interactions of amphetamine analogs with human liver CYP2D6. Biochem Pharmacol. 1997 Jun 1;53(11):1605-12. | ||||
| 411 | Identification of the human hepatic cytochromes P450 involved in the in vitro oxidation of antipyrine. Drug Metab Dispos. 1996 Apr;24(4):487-94. | ||||
| 412 | A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl Microbiol Biotechnol. 2010 Mar;86(1):163-75. | ||||
| 413 | Study on the cytochrome P450-mediated oxidative metabolism of the terpene alcohol linalool: indication of biological epoxidation | ||||
| 414 | Metabolism of geraniol and linalool in the rat and effects on liver and lung microsomal enzymes | ||||
| 415 | Metabolism of methoxyphenamine and 2-methoxyamphetamine in P4502D6-transfected cells and cell preparations. Xenobiotica. 1995 Sep;25(9):895-906. | ||||
| 416 | Oxidation of histamine H1 antagonist mequitazine is catalyzed by cytochrome P450 2D6 in human liver microsomes. J Pharmacol Exp Ther. 1998 Feb;284(2):437-42. | ||||
| 417 | Psychedelic 5-methoxy-N,N-dimethyltryptamine: metabolism, pharmacokinetics, drug interactions, and pharmacological actions. Curr Drug Metab. 2010 Oct;11(8):659-66. | ||||
| 418 | [Trazodon--the antidepressant: mechanism of action and its position in the treatment of depression]. Psychiatr Pol. 2011 Jul-Aug;45(4):611-25. | ||||
| 419 | Evaluation of interethnic differences in repinotan pharmacokinetics by using population approach. Drug Metab Pharmacokinet. 2006 Feb;21(1):61-9. | ||||
| 420 | Cytochrome P450 inactivation by pharmaceuticals and phytochemicals: therapeutic relevance Drug Metab Rev. 2008;40(1):101-47. doi: 10.1080/03602530701836704. | ||||
| 421 | Acetaminophen, via its reactive metabolite N-acetyl-p-benzo-quinoneimine and transient receptor potential ankyrin-1 stimulation, causes neurogenic inflammation in the airways and other tissues in rodents FASEB J. 2010 Dec;24(12):4904-16. doi: 10.1096/fj.10-162438. | ||||
| 422 | Rapid detection and quantification of paracetamol and its major metabolites using surface enhanced Raman scattering. Analyst. 2023 Apr 11;148(8):1805-1814. doi: 10.1039/d3an00249g. | ||||
| 423 | DrugBank(Pharmacology-Metabolism):Almogran | ||||
| 424 | Cytochromes P450: a structure-based summary of biotransformations using representative substrates Drug Metab Rev. 2008;40(1):1-100. doi: 10.1080/03602530802309742. | ||||
| 425 | CYP2D6 and CYP2C19 genotypes and amitriptyline metabolite ratios in a series of medicolegal autopsies | ||||
| 426 | Amitriptyline Therapy and CYP2D6 and CYP2C19 Genotype | ||||
| 427 | Addition of amoxapine improves positive and negative symptoms in a patient with schizophrenia. Ther Adv Psychopharmacol. 2013 Dec;3(6):340-2. | ||||
| 428 | DrugBank(Pharmacology-Metabolism)Amoxapine | ||||
| 429 | A simplified procedure for the analysis of formoterol in human urine by liquid chromatography-electrospray tandem mass spectrometry: application to the characterization of the metabolic profile and stability of formoterol in urine | ||||
| 430 | Mass balance and metabolism of [(3)H]Formoterol in healthy men after combined i.v. and oral administration-mimicking inhalation Drug Metab Dispos. 1999 Oct;27(10):1104-16. | ||||
| 431 | Current pharmacogenomic recommendations in chronic respiratory diseases: Is there a biomarker ready for clinical implementation? | ||||
| 432 | Aripiprazole Lauroxil: Pharmacokinetic Profile of This Long-Acting Injectable Antipsychotic in Persons With Schizophrenia J Clin Psychopharmacol. 2017 Jun;37(3):289-295. doi: 10.1097/JCP.0000000000000691. | ||||
| 433 | Biological conversion of?aripiprazole lauroxil - An N-acyloxymethyl aripiprazole prodrug Results Pharma Sci. 2014 May 2;4:19-25. doi: 10.1016/j.rinphs.2014.04.002. | ||||
| 434 | Effects of arotinolol, an alpha- and beta-adrenoceptor antagonist, on renin release from rat kidney cortical slices | ||||
| 435 | Metabolism of atenolol in man. Xenobiotica. 1978 May;8(5):313-20. | ||||
| 436 | DrugBank : Atenolol | ||||
| 437 | Disposition and metabolic fate of atomoxetine hydrochloride: the role of CYP2D6 in human disposition and metabolism | ||||
| 438 | The metabolism of berberine and its contribution to the pharmacological effects | ||||
| 439 | Transformation of berberine to its demethylated metabolites by the CYP51 enzyme in the gut microbiota | ||||
| 440 | Stereoselective metabolism of bisoprolol enantiomers in dogs and humans. Life Sci. 1998;63(13):1097-108. | ||||
| 441 | Glucuronidation as a major metabolic clearance pathway of 14c-labeled muraglitazar in humans: metabolic profiles in subjects with or without bile collection Drug Metab Dispos. 2006 Mar;34(3):427-39. doi: 10.1124/dmd.105.007617. | ||||
| 442 | Metabolism and disposition of [14C]BMS-690514, an ErbB/vascular endothelial growth factor receptor inhibitor, after oral administration to humans | ||||
| 443 | DrugBank(Pharmacology-Metabolism):Deucravacitinib | ||||
| 444 | U. S. FDA Label -BNP-1350 | ||||
| 445 | Metabolic pathway of icotinib in vitro: the differential roles of CYP3A4, CYP3A5, and CYP1A2 on potential pharmacokinetic drug-drug interaction. J Pharm Sci. 2018 Apr;107(4):979-983. | ||||
| 446 | Pharmacokinetics and metabolism of brexpiprazole, a novel serotonin-dopamine activity modulator and its main metabolite in rat, monkey and human. Xenobiotica. 2021 May;51(5):590-604. doi: 10.1080/00498254.2021.1890275. | ||||
| 447 | Metabolite profiling and reaction phenotyping for the in vitro assessment of the bioactivation of bromfenac. Chem Res Toxicol. 2020 Jan 21;33(1):249-257. | ||||
| 448 | Metabolic disposition of 14C-bromfenac in healthy male volunteers J Clin Pharmacol. 1998 Aug;38(8):744-52. doi: 10.1002/j.1552-4604.1998.tb04815.x. | ||||
| 449 | 4-Hydroxylation of debrisoquine by human CYP1A1 and its inhibition by quinidine and quinine. J Pharmacol Exp Ther. 2002 Jun;301(3):1025-32. | ||||
| 450 | In Vitro Interaction of AB-FUBINACA with Human Cytochrome P450, UDP-Glucuronosyltransferase Enzymes and Drug Transporters | ||||
| 451 | Structural elucidation of phase I and II metabolites of bupivacaine in horse urine and fungi of the Cunninghamella species using liquid chromatography/multi-stage mass spectrometry Rapid Commun Mass Spectrom. 2012 Jun 15;26(11):1338-46. doi: 10.1002/rcm.6225. | ||||
| 452 | Interactions of bupranolol with the polymorphic debrisoquine/sparteine monooxygenase (CYP2D6). Eur J Clin Pharmacol. 1993;45(3):261-4. | ||||
| 453 | DrugBank(Pharmacology-Metabolism)Buspirone | ||||
| 454 | Metabolism of butyrylfentanyl in fresh human hepatocytes: chemical synthesis of authentic metabolite standards for definitive identification. Biol Pharm Bull. 2019;42(4):623-630. | ||||
| 455 | Benzydamine N-oxygenation as an index for flavin-containing monooxygenase activity and benzydamine N-demethylation by cytochrome P450 enzymes in liver microsomes from rats, dogs, monkeys, and humans | ||||
| 456 | Metabolism of carvedilol in dogs, rats, and mice Drug Metab Dispos. 1998 Oct;26(10):958-69. | ||||
| 457 | The bacterial P450 BM3: a prototype for a biocatalyst with human P450 activities. Trends Biotechnol. 2007 Jul;25(7):289-98. | ||||
| 458 | Celecoxib pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics. 2012 Apr;22(4):310-8. | ||||
| 459 | Identification of human drug-metabolizing enzymes involved in the metabolism of SNI-2011. Biol Pharm Bull. 2001 Nov;24(11):1263-6. | ||||
| 460 | Determination of the new monoamine oxidase inhibitor brofaromine and its major metabolite in biological material by gas chromatography with electron-capture detection | ||||
| 461 | Clinical pharmacokinetics and metabolism of chloroquine. Focus on recent advancements Clin Pharmacokinet. 1996 Oct;31(4):257-74. doi: 10.2165/00003088-199631040-00003. | ||||
| 462 | Cytochrome P450 2C8 and CYP3A4/5 are involved in chloroquine metabolism in human liver microsomes. Arch Pharm Res. 2003 Aug;26(8):631-7. | ||||
| 463 | Metabolism of chlorpheniramine in rat and human by use of stable isotopes Xenobiotica. 1991 Jan;21(1):97-109. doi: 10.3109/00498259109039454. | ||||
| 464 | Classics in Chemical Neuroscience: Chlorpromazine ACS Chem Neurosci. 2019 Jan 16;10(1):79-88. doi: 10.1021/acschemneuro.8b00258. | ||||
| 465 | Determination of chlorpromazine and its metabolites in animal-derived foods using QuEChERS-based extraction, EMR-Lipid cleanup, and UHPLC-Q-Orbitrap MS analysis. Food Chem. 2023 Mar 1;403:134298. doi: 10.1016/j.foodchem.2022.134298. | ||||
| 466 | In Vitro metabolism of ciclesonide in human lung and liver precision-cut tissue slices Biopharm Drug Dispos. 2006 May;27(4):197-207. doi: 10.1002/bdd.500. | ||||
| 467 | PharmGKB summary: citalopram pharmacokinetics pathway. Pharmacogenet Genomics. 2011 Nov;21(11):769-72. | ||||
| 468 | Analytical methodologies for the enantiodetermination of citalopram and its metabolites | ||||
| 469 | Metabolism, pharmacokinetics, and excretion of a nonpeptidic substance P receptor antagonist, ezlopitant, in normal healthy male volunteers: characterization of polar metabolites by chemical derivatization with dansyl chloride | ||||
| 470 | Determination of clomipramine or imipramine and their mono-demethylated metabolites in human blood or plasma by high-performance liquid chromatography | ||||
| 471 | Metabolism of clomipramine in a Japanese psychiatric population: hydroxylation, desmethylation, and glucuronidation | ||||
| 472 | Biotransformation of cobicistat: metabolic pathways and enzymes. Drug Metab Lett. 2016;10(2):111-23. | ||||
| 473 | Pharmacogenetics of analgesic drugs | ||||
| 474 | Metabolism, pharmacokinetics, and excretion of the substance P receptor antagonist CP-122,721 in humans: structural characterization of the novel major circulating metabolite 5-trifluoromethoxy salicylic acid by high-performance liquid chromatography-tandem mass spectrometry and NMR spectroscopy | ||||
| 475 | Dacomitinib: an investigational drug for the treatment of glioblastoma Expert Opin Investig Drugs. 2018 Oct;27(10):823-829. doi: 10.1080/13543784.2018.1528225. | ||||
| 476 | In vitro characterization and pharmacokinetics of dapagliflozin (BMS-512148), a potent sodium-glucose cotransporter type II inhibitor, in animals and humans Drug Metab Dispos. 2010 Mar;38(3):405-14. doi: 10.1124/dmd.109.029165. | ||||
| 477 | Characterization of Phase I Hepatic Metabolites of Anti-Premature Ejaculation Drug Dapoxetine by UHPLC-ESI-Q-TOF | ||||
| 478 | Effects of Evodiamine on the Pharmacokinetics of Dapoxetine and Its Metabolite Desmethyl Dapoxetine in Rats | ||||
| 479 | Effects of 22 novel CYP2D6 variants found in Chinese population on the metabolism of dapoxetine | ||||
| 480 | Kinetics of deamination of 5-aza-2'-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside Biochem Pharmacol. 1983 Apr 1;32(7):1327-8. doi: 10.1016/0006-2952(83)90293-9. | ||||
| 481 | Pharmacokinetics and metabolism of delamanid, a novel anti-tuberculosis drug, in animals and humans: importance of albumin metabolism in vivo. Drug Metab Dispos. 2015 Aug;43(8):1267-76. | ||||
| 482 | Clinical pharmacokinetics of imipramine and desipramine | ||||
| 483 | Pharmacokinetic and Metabolic Profile of Deutetrabenazine (TEV-50717) Compared With Tetrabenazine in Healthy Volunteers Clin Transl Sci. 2020 Jul;13(4):707-717. doi: 10.1111/cts.12754. | ||||
| 484 | Clinical pharmacokinetics of amfetamine and related substances: monitoring in conventional and non-conventional matrices Clin Pharmacokinet. 2004;43(3):157-85. doi: 10.2165/00003088-200443030-00002. | ||||
| 485 | LC-MS/MS analysis of dextromethorphan metabolism in human saliva and urine to determine CYP2D6 phenotype and individual variability in N-demethylation and glucuronidation J Chromatogr B Analyt Technol Biomed Life Sci. 2004 Dec 25;813(1-2):217-25. doi: 10.1016/j.jchromb.2004.09.040. | ||||
| 486 | Classics in Chemical Neuroscience: Dextromethorphan (DXM). ACS Chem Neurosci. 2023 Jun 21;14(12):2256-2270. doi: 10.1021/acschemneuro.3c00088. | ||||
| 487 | CYP3A4 mediates dextropropoxyphene N-demethylation to nordextropropoxyphene: human in vitro and in vivo studies and lack of CYP2D6 involvement. Xenobiotica. 2004 Oct;34(10):875-87. | ||||
| 488 | The role of CYP2D6 in primary and secondary oxidative metabolism of dextromethorphan: in vitro studies using human liver microsomes. Br J Clin Pharmacol. 1994 Sep;38(3):243-8. | ||||
| 489 | Pharmacokinetics of dihydrocodeine and its active metabolite after single and multiple oral dosing. Br J Clin Pharmacol. 1999 Sep;48(3):317-22. | ||||
| 490 | Dihydrocodeine: safety concerns | ||||
| 491 | Pharmacokinetics and metabolism of diltiazem in healthy males and females following a single oral dose Eur J Drug Metab Pharmacokinet. 1993 Apr-Jun;18(2):199-206. doi: 10.1007/BF03188796. | ||||
| 492 | Desacetyl-diltiazem displays severalfold higher affinity to CYP2D6 compared with CYP3A4. Drug Metab Dispos. 2002 Jan;30(1):1-3. | ||||
| 493 | Bioconcentration, metabolism and effects of diphenhydramine on behavioral and biochemical markers in crucian carp (Carassius auratus) Sci Total Environ. 2016 Feb 15;544:400-9. doi: 10.1016/j.scitotenv.2015.11.132. | ||||
| 494 | Non-invasive skin sampling detects systemically administered drugs in humans. PLoS One. 2022 Jul 26;17(7):e0271794. doi: 10.1371/journal.pone.0271794. | ||||
| 495 | DrugBank(Pharmacology-Metabolism):Doxepin hydrochloride | ||||
| 496 | CYP2C9 and CYP2C19 polymorphic forms are related to increased indisulam exposure and higher risk of severe hematologic toxicity. Clin Cancer Res. 2007 May 15;13(10):2970-6. | ||||
| 497 | #N/A | ||||
| 498 | Disposition, profiling and identification of emixustat and its metabolites in humans | ||||
| 499 | Oxidative metabolism of encainide: polymorphism, pharmacokinetics and clinical considerations | ||||
| 500 | Simultaneous measurement of epinastine and its metabolite, 9,13b-dehydroepinastine, in human plasma by a newly developed ultra-performance liquid chromatography-tandem mass spectrometry and its application to pharmacokinetic studies Biomed Chromatogr. 2020 Sep;34(9):e4848. doi: 10.1002/bmc.4848. | ||||
| 501 | Metabolism and disposition in rats, dogs, and humans of erdafitinib, an orally administered potent pan-fibroblast growth factor receptor (FGFR) tyrosine kinase inhibitor Xenobiotica. 2021 Feb;51(2):177-193. doi: 10.1080/00498254.2020.1821123. | ||||
| 502 | Escitalopram (S-citalopram) and its metabolites in vitro: cytochromes mediating biotransformation, inhibitory effects, and comparison to R-citalopram Drug Metab Dispos. 2001 Aug;29(8):1102-9. | ||||
| 503 | Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003 Aug;144(8):3382-98. | ||||
| 504 | Aniline and its metabolites generate free radicals in yeast | ||||
| 505 | Absorption, metabolism, and excretion of etoricoxib, a potent and selective cyclooxygenase-2 inhibitor, in healthy male volunteers | ||||
| 506 | U. S. FDA Label -Fenfluramine | ||||
| 507 | Physiologically Based Pharmacokinetic Modeling Suggests Limited Drug-Drug Interaction for Fesoterodine When Coadministered With Mirabegron J Clin Pharmacol. 2019 Nov;59(11):1505-1518. doi: 10.1002/jcph.1438. | ||||
| 508 | LABEL:FESOTERODINE FUMARATE tablet, extended release | ||||
| 509 | Identification and characterization of in vitro and in vivo fidarestat metabolites: Toxicity and efficacy evaluation of metabolites | ||||
| 510 | Effects of CYP2D6 genotypes on age-related change of flecainide metabolism: involvement of CYP1A2-mediated metabolism. Br J Clin Pharmacol. 2009 Jul;68(1):89-96. | ||||
| 511 | Flibanserin therapy and CYP2C19 genotype. | ||||
| 512 | Simultaneous identification and quantitation of fluoxetine and its metabolite, norfluoxetine, in biological samples by GC-MS | ||||
| 513 | Roles of different CYP enzymes in the formation of specific fluvastatin metabolites by human liver microsomes. Basic Clin Pharmacol Toxicol. 2009 Nov;105(5):327-32. | ||||
| 514 | Metabolism and transport of oxazaphosphorines and the clinical implications Drug Metab Rev. 2005;37(4):611-703. doi: 10.1080/03602530500364023. | ||||
| 515 | Assessment of ifosfamide pharmacokinetics, toxicity, and relation to CYP3A4 activity as measured by the erythromycin breath test in patients with sarcoma Cancer. 2007 Jun 1;109(11):2315-22. doi: 10.1002/cncr.22669. | ||||
| 516 | Esophageal bezoar due to karaya gum granules used as a laxative | ||||
| 517 | Incarceration of the medial collateral ligament in the intercondylar notch following proximal avulsion | ||||
| 518 | A simple and rapid infrared-assisted self enzymolysis extraction method for total flavonoid aglycones extraction from Scutellariae Radix and mechanism exploration | ||||
| 519 | Management of postoperative junctional ectopic tachycardia in pediatric patients: a survey of 30 centers in Germany, Austria, and Switzerland | ||||
| 520 | Identification and determination of opipramol metabolites in plasma and urine | ||||
| 521 | The metabolism and excretion of galantamine in rats, dogs, and humans Drug Metab Dispos. 2002 May;30(5):553-63. doi: 10.1124/dmd.30.5.553. | ||||
| 522 | Ganaxolone | ||||
| 523 | Contribution of cytochrome P450 isoforms to gliquidone metabolism in rats and human | ||||
| 524 | Pharmacology, toxicology and clinical safety of glycopyrrolate | ||||
| 525 | Main contribution of the cytochrome P450 isoenzyme 1A2 (CYP1A2) to N-demethylation and 5-sulfoxidation of the phenothiazine neuroleptic chlorpromazine in human liver--A comparison with other phenothiazines. Biochem Pharmacol. 2010 Oct 15;80(8):1252-9. | ||||
| 526 | Update on hydrocodone metabolites in rats and dogs aided with a semi-automatic software for metabolite identification Mass-MetaSite | ||||
| 527 | DrugBank(Pharmacology-Metabolism):Hydrocodone bitartrate | ||||
| 528 | The single dose kinetics of chloroquine and its major metabolite desethylchloroquine in healthy subjects | ||||
| 529 | Iloperidone: chemistry, pharmacodynamics, pharmacokinetics and metabolism, clinical efficacy, safety and tolerability, regulatory affairs, and an opinion. Expert Opin Drug Metab Toxicol. 2010 Dec;6(12):1551-64. | ||||
| 530 | Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism | ||||
| 531 | Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes | ||||
| 532 | In vitro biotransformation of imatinib by the tumor expressed CYP1A1 and CYP1B1 | ||||
| 533 | The Use of In Vitro Data and Physiologically-Based Pharmacokinetic Modeling to Predict Drug Metabolite Exposure: Desipramine Exposure in Cytochrome P4502D6 Extensive and Poor Metabolizers Following Administration of Imipramine | ||||
| 534 | Comparative Pharmacokinetics and Safety of Imrecoxib, a Novel Selective Cyclooxygenase-2 Inhibitor, in Elderly Healthy Subjects. Drug Des Devel Ther. 2022 Nov 8;16:3865-3876. doi: 10.2147/DDDT.S387508. | ||||
| 535 | In vitro metabolism of indiplon and an assessment of its drug interaction potential | ||||
| 536 | Biotransformation of the ipecac alkaloids cephaeline and emetine from ipecac syrup in rats Eur J Drug Metab Pharmacokinet. 2002 Jan-Mar;27(1):29-35. doi: 10.1007/BF03190402. | ||||
| 537 | The role of levomilnacipran in the management of major depressive disorder: a comprehensive review. Curr Neuropharmacol. 2016;14(2):191-9. | ||||
| 538 | Advances in high-resolution MS and hepatocyte models solve a long-standing metabolism challenge: the loratadine story. Bioanalysis. 2016 Aug;8(16):1645-62. | ||||
| 539 | In vitro metabolism of perospirone in rat, monkey and human liver microsomes | ||||
| 540 | The wide variability of perospirone metabolism and the effect of perospirone on prolactin in psychiatric patients | ||||
| 541 | Sex- and dose-dependency in the pharmacokinetics and pharmacodynamics of (+)-methamphetamine and its metabolite (+)-amphetamine in rats | ||||
| 542 | Drugs of abuse in a non-conventional sample; detection of methamphetamine and its main metabolite, amphetamine in abusers' clothes by HPLC with UV and fluorescence detection | ||||
| 543 | Reduced CYP2D6 activity is a negative risk factor for methamphetamine dependence | ||||
| 544 | Effects of methamphetamine and its primary human metabolite, p-hydroxymethamphetamine, on the development of the Australian blowfly Calliphora stygia | ||||
| 545 | Effects of cytochrome P450 single nucleotide polymorphisms on methadone metabolism and pharmacodynamics | ||||
| 546 | Interindividual and interspecies variation in the metabolism of the hallucinogen 4-methoxyamphetamine | ||||
| 547 | Identification of novel metoclopramide metabolites in humans: in vitro and in vivo studies | ||||
| 548 | The gastroprokinetic and antiemetic drug metoclopramide is a substrate and inhibitor of cytochrome P450 2D6. Drug Metab Dispos. 2002 Mar;30(3):336-43. | ||||
| 549 | Lessons from pharmacogenetics and metoclopramide: toward the right dose of the right drug for the right patient. J Clin Gastroenterol. 2012 Jul;46(6):437-9. | ||||
| 550 | DrugBank(Pharmacology-Metabolism):Metoprolol succinate | ||||
| 551 | Metabolism of metoprolol in the rat in vitro and in vivo | ||||
| 552 | Involvement of CYP1A2 in mexiletine metabolism. Br J Clin Pharmacol. 1998 Jul;46(1):55-62. | ||||
| 553 | The in vitro metabolism of desglymidodrine, an active metabolite of prodrug midodrine by human liver microsomes | ||||
| 554 | The biotransformation of [14C]minaprine in man and five animals species Xenobiotica. 1981 Nov;11(11):735-47. doi: 10.3109/00498258109045877. | ||||
| 555 | Clinical pharmacokinetics of mirtazapine | ||||
| 556 | The pharmacokinetics of bucindolol and its major metabolite in essential hypertension | ||||
| 557 | Population pharmacokinetic modeling of motesanib and its active metabolite, M4, in cancer patients | ||||
| 558 | DrugBank(Pharmacology-Metabolism):Adagrasib | ||||
| 559 | Identification of human cytochrome P450 isozymes involved in the metabolism of naftopidil enantiomers in vitro. J Pharm Pharmacol. 2014 Nov;66(11):1534-51. | ||||
| 560 | Characterization of Ca(2+)-antagonistic effects of three metabolites of the new antihypertensive agent naftopidil, (naphthyl)hydroxy-naftopidil, (phenyl)hydroxy-naftopidil, and O-desmethyl-naftopidil J Cardiovasc Pharmacol. 1991 Dec;18(6):918-25. doi: 10.1097/00005344-199112000-00020. | ||||
| 561 | Evaluation of 24 CYP2D6 variants on the metabolism of nebivolol in vitro. Drug Metab Dispos. 2016 Nov;44(11):1828-1831. | ||||
| 562 | Absorption, disposition, metabolic fate, and elimination of the dopamine agonist rotigotine in man: administration by intravenous infusion or transdermal delivery | ||||
| 563 | Disposition and biotransformation of the antiretroviral drug nevirapine in humans Drug Metab Dispos. 1999 Aug;27(8):895-901. | ||||
| 564 | Differences in nevirapine biotransformation as a factor for its sex-dependent dimorphic profile of adverse drug reactions J Antimicrob Chemother. 2014 Feb;69(2):476-82. doi: 10.1093/jac/dkt359. | ||||
| 565 | Sulfation of 12-hydroxy-nevirapine by human SULTs and the effects of genetic polymorphisms of SULT1A1 and SULT2A1. Biochem Pharmacol. 2022 Oct;204:115243. doi: 10.1016/j.bcp.2022.115243. | ||||
| 566 | U. S. FDA Label -Nevirapine | ||||
| 567 | Assay of nicergoline and three metabolites in human plasma and urine by high-performance liquid chromatography-atmospheric pressure ionization mass spectrometry J Chromatogr. 1991 Aug 23;568(2):375-84. doi: 10.1016/0378-4347(91)80175-c. | ||||
| 568 | Pharmacokinetics of nortriptyline and its 10-hydroxy metabolite in Chinese subjects of different CYP2D6 genotypes Clin Pharmacol Ther. 1998 Oct;64(4):384-90. doi: 10.1016/S0009-9236(98)90069-8. | ||||
| 569 | Quantitative Modeling Analysis Demonstrates the Impact of CYP2C19 and CYP2D6 Genetic Polymorphisms on the Pharmacokinetics of Amitriptyline and Its Metabolite, Nortriptyline J Clin Pharmacol. 2019 Apr;59(4):532-540. doi: 10.1002/jcph.1344. | ||||
| 570 | DrugBank(Pharmacology-Metabolism)Nortriptyline hydrochloride | ||||
| 571 | Olanzapine. Pharmacokinetic and pharmacodynamic profile. Clin Pharmacokinet. 1999 Sep;37(3):177-93. | ||||
| 572 | The Unique Human N10-Glucuronidated Metabolite Formation from Olanzapine in Chimeric NOG-TKm30 Mice with Humanized Livers. Drug Metab Dispos. 2023 Apr;51(4):480-491. doi: 10.1124/dmd.122.001102. | ||||
| 573 | Quantification of olanzapine and its three metabolites by liquid chromatography-tandem mass spectrometry in human body fluids obtained from four deceased, and confirmation of the reduction from olanzapine N-oxide to olanzapine in whole blood in vitro. Forensic Toxicol. 2023 Jul;41(2):318-328. doi: 10.1007/s11419-023-00662-0. | ||||
| 574 | Identification of novel ondansetron metabolites using LC/MS(n) and NMR J Chromatogr B Analyt Technol Biomed Life Sci. 2018 Sep 15;1095:138-141. doi: 10.1016/j.jchromb.2018.07.013. | ||||
| 575 | Human cytochromes mediating gepirone biotransformation at low substrate concentrations. Biopharm Drug Dispos. 2003 Mar;24(2):87-94. | ||||
| 576 | Metabolism and Disposition of Prescription Opioids: A Review | ||||
| 577 | Zhx2 Is a Candidate Gene Underlying Oxymorphone Metabolite Brain Concentration Associated with State-Dependent Oxycodone Reward. J Pharmacol Exp Ther. 2022 Aug;382(2):167-180. doi: 10.1124/jpet.122.001217. | ||||
| 578 | Impact of genetic variation in CYP2C19, CYP2D6, and CYP3A4 on oxycodone and its metabolites in a large database of clinical urine drug tests. Pharmacogenomics J. 2022 Feb;22(1):25-32. doi: 10.1038/s41397-021-00253-5. | ||||
| 579 | Association of CYP2D6 genotype predicted phenotypes with oxycodone requirements and side effects in children undergoing surgery. Ann Transl Med. 2022 Dec;10(23):1262. doi: 10.21037/atm-2022-58. | ||||
| 580 | Different enantioselective 9-hydroxylation of risperidone by the two human CYP2D6 and CYP3A4 enzymes. Drug Metab Dispos. 2001 Oct;29(10):1263-8. | ||||
| 581 | S73 | METXBIODB | metabolite Reaction Database from BioTransformer | DOI:10.5281/zenodo.4056560 | ||||
| 582 | Pharmacokinetics, metabolism and excretion of intravenous [l4C]-palonosetron in healthy human volunteers Biopharm Drug Dispos. 2004 Nov;25(8):329-37. doi: 10.1002/bdd.410. | ||||
| 583 | Identification of cytochrome P450 isoforms involved in the metabolism of paroxetine and estimation of their importance for human paroxetine metabolism using a population-based simulator. Drug Metab Dispos. 2010 Mar;38(3):376-85. | ||||
| 584 | Pazopanib, a new therapy for metastatic soft tissue sarcoma. Expert Opin Pharmacother. 2013 May;14(7):929-35. | ||||
| 585 | Bioavailability, metabolism and disposition of oral pazopanib in patients with advanced cancer Xenobiotica. 2013 May;43(5):443-53. doi: 10.3109/00498254.2012.734642. | ||||
| 586 | Anti-Parkinson's disease drugs and pharmacogenetic considerations Expert Opin Drug Metab Toxicol. 2013 Jul;9(7):859-74. doi: 10.1517/17425255.2013.789018. | ||||
| 587 | DrugBank(Pharmacology-Metabolism)Phenacetin | ||||
| 588 | The Respective Roles of CYP3A4 and CYP2D6 in the Metabolism of Pimozide to Established and Novel Metabolites Drug Metab Dispos. 2020 Nov;48(11):1113-1120. doi: 10.1124/dmd.120.000188. | ||||
| 589 | Hazardous Substances Data Bank:Piperazine | ||||
| 590 | Metabolic fate of pitavastatin, a new inhibitor of HMG-CoA reductase: similarities and difference in the metabolism of pitavastatin in monkeys and humans | ||||
| 591 | FDA Center for Drug Evaluation and Research: Pitolisant Non-clinical Review(s) | ||||
| 592 | Wakix, INN-Pitolisant - European Medicines Agency - Europa EU | ||||
| 593 | Absorption, metabolism, and excretion of [(14)C]ponatinib after a single oral dose in humans Cancer Chemother Pharmacol. 2017 Mar;79(3):507-518. doi: 10.1007/s00280-017-3240-x. | ||||
| 594 | In vitro metabolites characterization of ponatinib in human liver microsomes using ultra-high performance liquid chromatography combined with Q-Exactive-Orbitrap tandem mass spectrometry Biomed Chromatogr. 2020 Jun;34(6):e4819. doi: 10.1002/bmc.4819. | ||||
| 595 | Quantitative analysis of primaquine and its metabolites in human urine using liquid chromatography coupled with tandem mass spectrometry J Chromatogr B Analyt Technol Biomed Life Sci. 2022 Dec 15;1213:123517. doi: 10.1016/j.jchromb.2022.123517. | ||||
| 596 | Enantioselective metabolism of primaquine by human CYP2D6 Malar J. 2014 Dec 17;13:507. doi: 10.1186/1475-2875-13-507. | ||||
| 597 | Metabolites of procainamide and practolol inhibit complement components C3 and C4 Biochem J. 1988 Apr 15;251(2):323-6. doi: 10.1042/bj2510323. | ||||
| 598 | Influence of cytochrome P450 genotype on the plasma disposition of prochlorperazine metabolites and their relationships with clinical responses in cancer patients Ann Clin Biochem. 2018 May;55(3):385-393. doi: 10.1177/0004563217731432. | ||||
| 599 | Simultaneous determination of prochlorperazine and its metabolites in human plasma using isocratic liquid chromatography tandem mass spectrometry Biomed Chromatogr. 2012 Jun;26(6):754-60. doi: 10.1002/bmc.1725. | ||||
| 600 | Bioavailability and metabolism of prochlorperazine administered via the buccal and oral delivery route J Clin Pharmacol. 2005 Dec;45(12):1383-90. doi: 10.1177/0091270005281044. | ||||
| 601 | Progesterone and testosterone hydroxylation by cytochromes P450 2C19, 2C9, and 3A4 in human liver microsomes. Arch Biochem Biophys. 1997 Oct 1;346(1):161-9. | ||||
| 602 | An HPLC-ESI-MS method for simultaneous determination of fourteen metabolites of promethazine and caffeine and its application to pharmacokinetic study of the combination therapy against motion sickness J Pharm Biomed Anal. 2012 Mar 25;62:119-28. doi: 10.1016/j.jpba.2011.12.033. | ||||
| 603 | A liquid chromatographic method for the simultaneous determination of promethazine and three of its metabolites in plasma using electrochemical and UV detectors J Chromatogr Sci. 2001 Feb;39(2):70-2. doi: 10.1093/chromsci/39.2.70. | ||||
| 604 | CYP2D6 is the principal cytochrome P450 responsible for metabolism of the histamine H1 antagonist promethazine in human liver microsomes. Pharmacogenetics. 1996 Oct;6(5):449-57. | ||||
| 605 | A rapid HPLC assay for the simultaneous determination of propafenone and its major metabolites in human serum Anal Sci. 2004 Sep;20(9):1307-11. doi: 10.2116/analsci.20.1307. | ||||
| 606 | Isolation, purification, and structure identification of glucuronic acid conjugates of propranolol and alprenolol and their ring-hydroxylated metabolites Drug Metab Dispos. 1984 Nov-Dec;12(6):749-54. | ||||
| 607 | Rapid and simultaneous extraction of propranolol, its neutral and basic metabolites from plasma and assay by high-performance liquid chromatography J Chromatogr. 1985 Oct 11;343(2):349-58. doi: 10.1016/s0378-4347(00)84603-4. | ||||
| 608 | Poncirin and its metabolite ponciretin attenuate colitis in mice by inhibiting LPS binding on TLR4 of macrophages and correcting Th17/Treg imbalance | ||||
| 609 | Metabolism of quetiapine by CYP3A4 and CYP3A5 in presence or absence of cytochrome B5. Drug Metab Dispos. 2009 Feb;37(2):254-8. | ||||
| 610 | Metabolism of the active metabolite of quetiapine, N-desalkylquetiapine in vitro. Drug Metab Dispos. 2012 Sep;40(9):1778-84. | ||||
| 611 | CYP450 phenotyping and metabolite identification of quinine by accurate mass UPLC-MS analysis: a possible metabolic link to blackwater fever | ||||
| 612 | Netupitant PET imaging and ADME studies in humans | ||||
| 613 | Ranolazine for the treatment of chronic angina and potential use in other cardiovascular conditions | ||||
| 614 | Fatal, Intentional Overdose of Ranolazine: Post-Mortem Distribution of Parent Drug and its Major Metabolite | ||||
| 615 | Comparison of pharmacokinetic parameters of ranolazine between diabetic and non-diabetic rats | ||||
| 616 | Effects of CYP3A Inhibition, CYP3A Induction, and Gastric Acid Reduction on the Pharmacokinetics of Ripretinib, a Switch Control KIT Tyrosine Kinase Inhibitor | ||||
| 617 | LABEL:Risperidone | ||||
| 618 | Biotransformation of moclobemide in humans | ||||
| 619 | Pharmacokinetics and safety of rucaparib in patients with advanced solid tumors and hepatic impairment. Cancer Chemother Pharmacol. 2021 Aug;88(2):259-270. doi: 10.1007/s00280-021-04278-2. | ||||
| 620 | Investigation of the metabolism of rufinamide and its interaction with valproate. Drug Metab Lett. 2011 Dec;5(4):280-9. | ||||
| 621 | A Phase 1 Study to Evaluate Absolute Bioavailability and Absorption, Distribution, Metabolism, and Excretion of Savolitinib in Healthy Male Volunteers | ||||
| 622 | Comparative studies on the cytochrome p450-associated metabolism and interaction potential of selegiline between human liver-derived in vitro systems. Drug Metab Dispos. 2003 Sep;31(9):1093-102. | ||||
| 623 | Metabolism of sertindole: identification of the metabolites in the rat and dog, and species comparison of liver microsomal metabolism | ||||
| 624 | Sertindole : a review of its use in schizophrenia | ||||
| 625 | Sertraline is metabolized by multiple cytochrome P450 enzymes, monoamine oxidases, and glucuronyl transferases in human: an in vitro study. Drug Metab Dispos. 2005 Feb;33(2):262-70. | ||||
| 626 | Pharmacogenetic Testing and Therapeutic Drug Monitoring Of Sertraline at a Residential Treatment Center for Children and Adolescents: A Pilot Study. Innov Pharm. 2022 Dec 26;13(4):10.24926/iip.v13i4.5035. doi: 10.24926/iip.v13i4.5035. | ||||
| 627 | Sertraline. 2022 May 15. Drugs and Lactation Database (LactMed?) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006C. | ||||
| 628 | Neonicotinoids and pharmaceuticals in hair of the Red fox (Vulpes vulpes) from the Cavallino-Treporti peninsula, Italy. Environ Res. 2023 Jul 1;228:115837. doi: 10.1016/j.envres.2023.115837. | ||||
| 629 | Functional Measurement of CYP2C9 and CYP3A4 Allelic Polymorphism on Sildenafil Metabolism | ||||
| 630 | Contribution of CYP3A isoforms to dealkylation of PDE5 inhibitors: a comparison between sildenafil N-demethylation and tadalafil demethylenation. Biol Pharm Bull. 2015;38(1):58-65. | ||||
| 631 | The contributions of cytochromes P450 3A4 and 3A5 to the metabolism of the phosphodiesterase type 5 inhibitors sildenafil, udenafil, and vardenafil. Drug Metab Dispos. 2008 Jun;36(6):986-90. | ||||
| 632 | Pharmacokinetics of sildenafil after single oral doses in healthy male subjects: absolute bioavailability, food effects and dose proportionality. Br J Clin Pharmacol. 2002;53 Suppl 1(Suppl 1):5S-12S. doi: 10.1046/j.0306-5251.2001.00027.x. | ||||
| 633 | Erectile dysfunction drugs changed the protein expressions and activities of drug-metabolising enzymes in the liver of male rats. Oxid Med Cell Longev. 2016;2016:4970906. | ||||
| 634 | Regioselectivity and stereoselectivity of the metabolism of the chiral quinolizidine alkaloids sparteine and pachycarpine in the rat | ||||
| 635 | Tafenoquine treatment of Plasmodium vivax malaria: suggestive evidence that CYP2D6 reduced metabolism is not associated with relapse in the Phase 2b DETECTIVE trial Malar J. 2016 Feb 18;15:97. doi: 10.1186/s12936-016-1145-5. | ||||
| 636 | PharmGKB:Tamoxifen citrate | ||||
| 637 | Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4'-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002 Aug;30(8):869-74. | ||||
| 638 | Endoxifen and other metabolites of tamoxifen inhibit human hydroxysteroid sulfotransferase 2A1 (hSULT2A1). Drug Metab Dispos. 2014 Nov;42(11):1843-50. | ||||
| 639 | Metabolism and transport of tamoxifen in relation to its effectiveness: new perspectives on an ongoing controversy. Future Oncol. 2014 Jan;10(1):107-22. | ||||
| 640 | Identification and biological activity of tamoxifen metabolites in human serum Biochem Pharmacol. 1983 Jul 1;32(13):2045-52. doi: 10.1016/0006-2952(83)90425-2. | ||||
| 641 | PharmGKB summary: tamoxifen pathway, pharmacokinetics Pharmacogenet Genomics. 2013 Nov;23(11):643-7. doi: 10.1097/FPC.0b013e3283656bc1. | ||||
| 642 | The tamoxifen dilemma. Carcinogenesis. 1999 Jul;20(7):1153-60. | ||||
| 643 | Pharmacokinetics and plasma protein binding of tamsulosin hydrochloride in rats, dogs, and humans Drug Metab Dispos. 1998 Mar;26(3):240-5. | ||||
| 644 | Absorption, metabolism and excretion of tamsulosin hydrochloride in man Xenobiotica. 1996 Jun;26(6):637-45. doi: 10.3109/00498259609046739. | ||||
| 645 | Simultaneous determination of tandospirone and its active metabolite, 1-[2-pyrimidyl]-piperazine in rat plasma by LC-MS/MS and its application to a pharmacokinetic study | ||||
| 646 | Clinical assessment of drug-drug interactions of tasimelteon, a novel dual melatonin receptor agonist. J Clin Pharmacol. 2015 Sep;55(9):1004-11. | ||||
| 647 | DrugBank(Pharmacology-Metabolism):Tetrabenazine | ||||
| 648 | Extraction of structure-activity relationship information from high-throughput screening data Curr Med Chem. 2009;16(31):4049-57. doi: 10.2174/092986709789378189. | ||||
| 649 | Timolol metabolism in human liver microsomes is mediated principally by CYP2D6. Drug Metab Dispos. 2007 Jul;35(7):1135-41. | ||||
| 650 | Metabolism of ophthalmic timolol: new aspects of an old drug. Basic Clin Pharmacol Toxicol. 2011 May;108(5):297-303. | ||||
| 651 | Quantitative determination of pentazocine in plasma and of pentazocine and metabolites in urine | ||||
| 652 | Metabolism-mediated drug interactions associated with ritonavir-boosted tipranavir in mice. Drug Metab Dispos. 2010 May;38(5):871-8. | ||||
| 653 | Trends in Tramadol: Pharmacology, Metabolism, and Misuse Anesth Analg. 2017 Jan;124(1):44-51. doi: 10.1213/ANE.0000000000001683. | ||||
| 654 | Simultaneous LC-MS/MS quantification of oxycodone, tramadol and fentanyl and their metabolites (noroxycodone, oxymorphone, O- desmethyltramadol, N- desmethyltramadol, and norfentanyl) in human plasma and whole blood collected via venepuncture and volumetric absorptive micro sampling. J Pharm Biomed Anal. 2021 Sep 5;203:114171. doi: 10.1016/j.jpba.2021.114171. | ||||
| 655 | Total determination of triclabendazole and its metabolites in bovine tissues using liquid chromatography-tandem mass spectrometry J Chromatogr B Analyt Technol Biomed Life Sci. 2019 Mar 1;1109:54-59. doi: 10.1016/j.jchromb.2019.01.021. | ||||
| 656 | Egaten FDA label | ||||
| 657 | In Vitro Hepatic Oxidative Biotransformation of Trimethoprim Drug Metab Dispos. 2015 Sep;43(9):1372-80. doi: 10.1124/dmd.115.065193. | ||||
| 658 | Characterization of the cytochrome P450 enzymes involved in the in vitro metabolism of dolasetron. Comparison with other indole-containing 5-HT3 antagonists. Drug Metab Dispos. 1996 May;24(5):602-9. | ||||
| 659 | The polymorphic cytochrome P-4502D6 is involved in the metabolism of both 5-hydroxytryptamine antagonists, tropisetron and ondansetron. Drug Metab Dispos. 1994 Mar-Apr;22(2):269-74. | ||||
| 660 | Pharmacokinetics and metabolism of the 5-hydroxytryptamine antagonist tropisetron after single oral doses in humans | ||||
| 661 | The Utilization of Mu-Opioid Receptor Biased Agonists: Oliceridine, an Opioid Analgesic with Reduced Adverse Effects | ||||
| 662 | CLINICAL PHARMACOLOGY AND BIOPHARMACEUTICS REVIEW: Umeclidinium Bromide | ||||
| 663 | LABEL:INVOQ- upadacitinib tablet, extended release | ||||
| 664 | O- and N-demethylation of venlafaxine in vitro by human liver microsomes and by microsomes from cDNA-transfected cells: effect of metabolic inhibitors and SSRI antidepressants. Neuropsychopharmacology. 1999 May;20(5):480-90. | ||||
| 665 | Venlafaxine oxidation in vitro is catalysed by CYP2D6. Br J Clin Pharmacol. 1996 Feb;41(2):149-56. | ||||
| 666 | Comparison of the pharmacokinetics of venlafaxine extended release and desvenlafaxine in extensive and poor cytochrome P450 2D6 metabolizers | ||||
| 667 | Accidental intoxications in toddlers: lack of cross-reactivity of vilazodone and its urinary metabolite M17 with drug of abuse screening immunoassays | ||||
| 668 | DETERMINATION OF VENLAFAXINE, VILAZODONE AND THEIR MAIN ACTIVE METABOLITES IN HUMAN SERUM BY HPLC-DAD AND HPLC-MS | ||||
| 669 | Vortioxetine: clinical pharmacokinetics and drug interactions. Clin Pharmacokinet. 2018 Jun;57(6):673-686. | ||||
| 670 | Metabolism of a potential 8-aminoquinoline antileishmanial drug in rat liver microsomes | ||||
| 671 | Zolpidem urine excretion profiles and cross-reactivity with ELISA(?) kits in subjects using Zolpidem or Ambien(?) CR as a prescription sleep aid | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Yin and Dr. Li.

